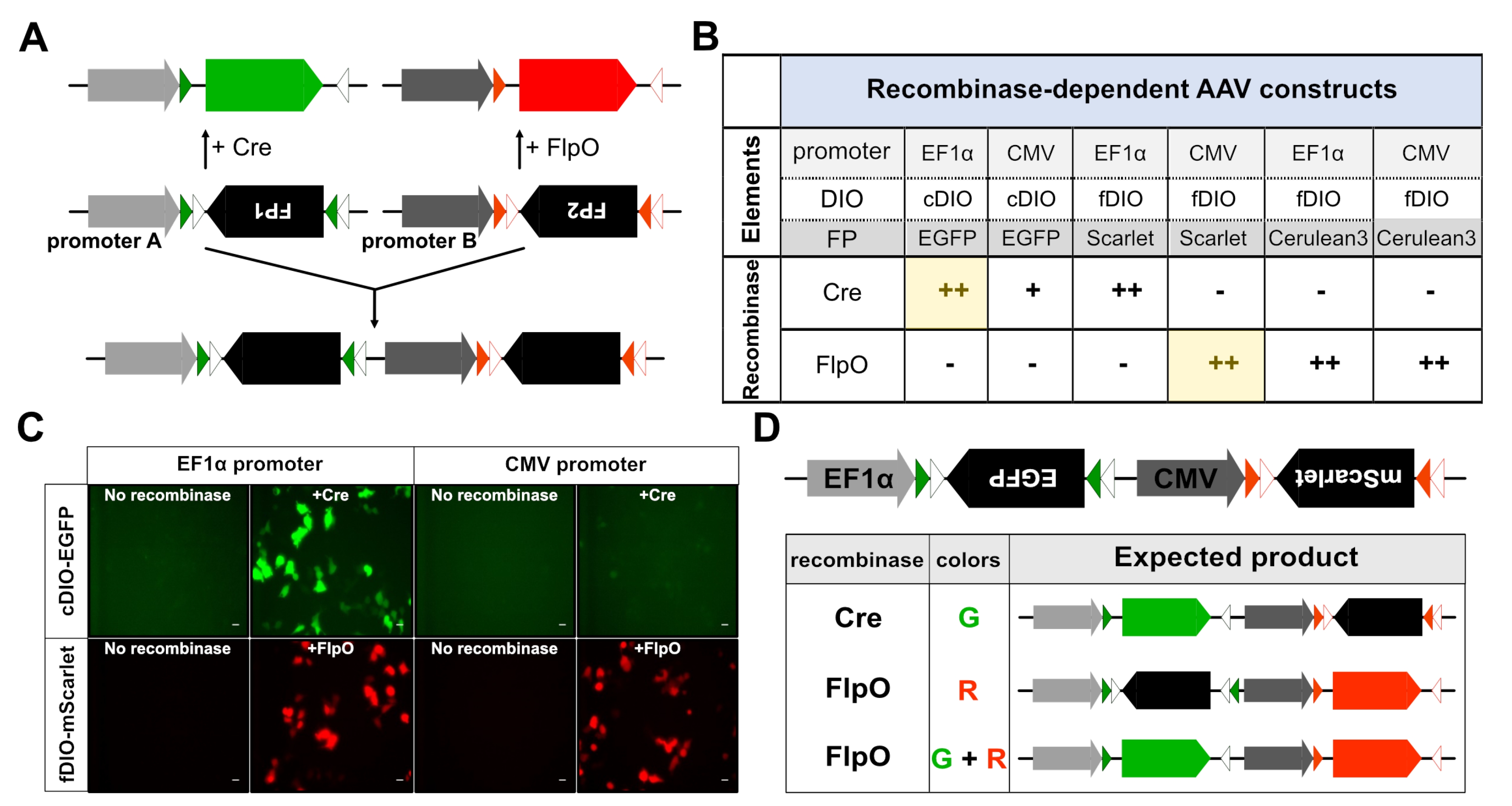
Fig. 2. Strategy for constructing dual color labeling vector. (A) Schematic diagram showing a dual-color labeling vector combining the Cre/loxP and FlpO/FRT recombination elements in the construct. The double-floxed inverse orientation (DIO), an inverted open reading frame (ORF) of fluorescent protein (FP) genes, is located between sets of recombinase recognition sites such as loxp/lox2272 (cDIO) or FRT/F5 (fDIO). The Cre- or FlpO-recombinase recognizes the cDIO (green/white triangles) or FRT/F5 (red/white triangles) sites, respectively, resulting in gene expression by inverting the ORF of the FPs through DNA recombination. cDIO-FP1 and fDIO-FP2 expression cassettes with promoters A and B were fused to produce our dual color labeling vector. (B) Summary of expression screening of recombinase-specific expression of AAV constructs. ++: high expression of FPs, +: moderate or weak expression of FPs, -: no expression of FPs. The combination with yellow color was selected to produce a fusion construct of EF1a-cDIO-EGFP-CMV-fDIO-mScarlet. (C) Representative fluorescence images showing recombinase-specific expression of fluorescent proteins (ex. EGFP and mScarlet). To screen for efficient and selective promoter-DIO-FP component pairs, HEK293T cells were co-transfected with promoter-DIO-FP vectors with or without vectors containing recombinase genes as indicated. In this figure, pAAV-cDIO or pAAV-fDIO vectors including EF1α or CMV promoter, and EGFP or mScarlet, were co-transfected with pAAV-Cre or pAAV-FlpO; scale bar: 20 μm. (D) Schematic diagram showing the expected FP expression from the dual color expression vector with the combined actions of Cre and FlpO recombinases.
© Exp Neurobiol


