-
Review Article | February 29, 2024
This article provides an review of autoimmune encephalitis (AIE) and its underlying pathological mechanisms. AIE is a rare condition characterized by inflammation in the brain parenchyma, often triggered by autoimmune attacks on specific neuronal epitopes. The article emphasizes the need for early diagnosis and effective treatment while highlighting the challenges in diagnosing AIE and the recognition of CD8+ lymphocytic encephalitis.
Show more This article provides an review of autoimmune encephalitis (AIE) and its underlying pathological mechanisms. AIE is a rare condition characterized by inflammation in the brain parenchyma, often triggered by autoimmune attacks on specific neuronal epitopes. The article emphasizes the need for early diagnosis and effective treatment while highlighting the challenges in diagnosing AIE and the recognition of CD8+ lymphocytic encephalitis.Yu-Mi Shim, Seong-Ik Kim, So Dug Lim et al.
This article provides an review of autoimmune encephalitis (AIE) and its underlying pathological mechanisms. AIE is a rare condition characterized by inflammation in the brain parenchyma, often triggered by autoimmune attacks on specific neuronal epitopes. The article emphasizes the need for early diagnosis and effective treatment while highlighting the challenges in diagnosing AIE and the recognition of CD8+ lymphocytic encephalitis.Yu-Mi Shim, Seong-Ik Kim, So Dug Lim et al. -
Original Article | February 29, 2024
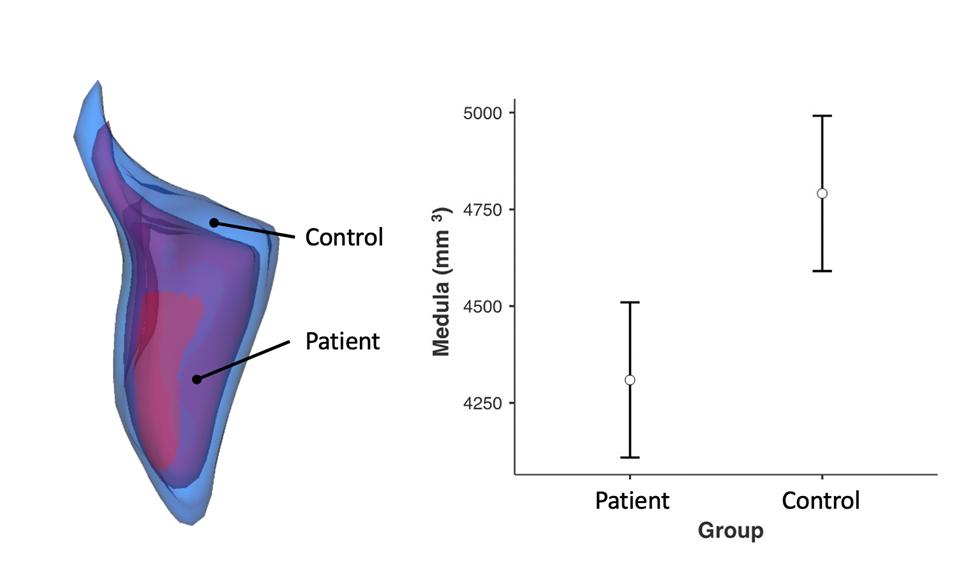
There was reduced medulla volume in patients with panic disorder. This may be related to panic disorder pathophysiology, given the medulla’s role in fear and autonomic function.
Show more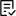 There was reduced medulla volume in patients with panic disorder. This may be related to panic disorder pathophysiology, given the medulla’s role in fear and autonomic function.Hye-Min Kim, Chanmi Kang, Boram Chae et al.
There was reduced medulla volume in patients with panic disorder. This may be related to panic disorder pathophysiology, given the medulla’s role in fear and autonomic function.Hye-Min Kim, Chanmi Kang, Boram Chae et al. -
Original Article | February 29, 2024
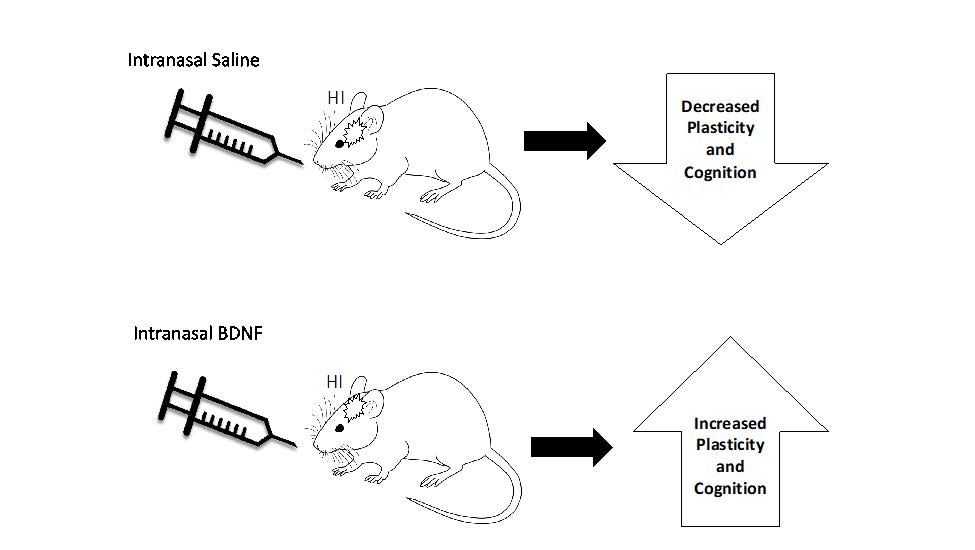
There is a need to explore noninvasive treatment after neonatal hypoxic ischemia. Intranasal administration of a 7-day BDNF treatment in PND 7 neonatal mice, resulted in cognitive rescue and neural plasticity.
Show more There is a need to explore noninvasive treatment after neonatal hypoxic ischemia. Intranasal administration of a 7-day BDNF treatment in PND 7 neonatal mice, resulted in cognitive rescue and neural plasticity.Serena-Kaye Sims, Madelynne Saddow, Lilly McGonegal and Catrina Sims-Robinson
There is a need to explore noninvasive treatment after neonatal hypoxic ischemia. Intranasal administration of a 7-day BDNF treatment in PND 7 neonatal mice, resulted in cognitive rescue and neural plasticity.Serena-Kaye Sims, Madelynne Saddow, Lilly McGonegal and Catrina Sims-Robinson -
Original Article | February 29, 2024
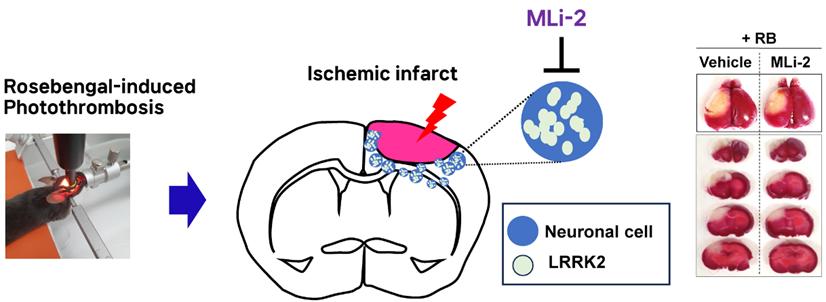
Endogenous LRRK2 was significantly increased in stroke models. LRRK2 inhibition has neuroprotective effects in that. LRRK2 targeted therapies could be helpful for treating stroke
Show more Endogenous LRRK2 was significantly increased in stroke models. LRRK2 inhibition has neuroprotective effects in that. LRRK2 targeted therapies could be helpful for treating strokeJeong-Ah Hwang, Seung Kyu Choi, Seong Hwan Kim and Dong Woon Kim
Endogenous LRRK2 was significantly increased in stroke models. LRRK2 inhibition has neuroprotective effects in that. LRRK2 targeted therapies could be helpful for treating strokeJeong-Ah Hwang, Seung Kyu Choi, Seong Hwan Kim and Dong Woon Kim
VISITORS
since Jan. 01, 2020| TODAY | 213 |
|---|---|
| TOTAL | 580,752 |
Highly Cited Papers
-
Review Article | December 30, 2015Geon Ha Kim, Jieun E. Kim, Sandy Jeong Rhie and Sujung Yoon
-
Review Article | March 30, 2013Onyou Hwang*
-
Review Article | June 30, 2015Hyo Eun Moon and Sun Ha Paek
News
-
한국과총에서 주관하는 「2023년 제33회 과학기술우수논문상」에 우리 학회 학술지 EN에 발간된 논문이 선정되었습니다.&n...…
Show more2023-06-23 -
Here is the winner of the 2021 EN Best Article Awards announced at the KSBNS 2021 Virtual Conference held on May 20~21, 2021. Congratulations to Dr. Joungha Won...…
Show more2021-05-28 -
Here are the winners of the 2020 EN Best Article Awards announced at the KSBNS 2020 Virtual Conference held onNovember 16~17, 2020. - Review Article Congratulat...…
Show more2020-11-18

 PDF
PDF