Articles
Article Tools
Stats or Metrics
Article
Original Article
Exp Neurobiol 2016; 25(3): 120-129
Published online June 30, 2016
https://doi.org/10.5607/en.2016.25.3.120
© The Korean Society for Brain and Neural Sciences
Neuronal Expression and Cell-Type-Specific Gene-Silencing of Best1 in Thalamic Reticular Nucleus Neurons Using pSico-Red System
Jae-Young Jung1,3, Seung Eun Lee2, Eun Mi Hwang1,3 and C. Justin Lee1,3,4*
1Center for Neuroscience and Functional Connectomics, Brain Science Institute, Korea Institute of Science and Technology (KIST), Seoul 02792, 2Research Animal Resource Center, Korea Institute of Science and Technology (KIST), Seoul 02792, 3Neuroscience Program, University of Science and Technology (UST), Daejeon 34113,4KU-KIST School of Converging Science and Technology, Korea University, Seoul 02841, Korea
Correspondence to: *To whom correspondence should be addressed.
TEL: 82-2-958-6940, FAX: 82-2-958-6919
e-mail: cjl@kist.re.kr
Assessing the cell-type expression pattern of a certain gene can be achieved by using cell-type-specific gene manipulation. Recently, cre-recombinase-dependent gene-silencing tool, pSico has become popular in neuroscientific research. However, pSico has a critical limitation that gene-silenced cell cannot be identified by fluorescence, due to an excision of the reporter gene for green fluorescence protein (GFP). To overcome this limitation, we newly developed pSico-Red, with
Keywords: bestrophin-1, thalamic reticular nucleus, pSico, virus, DLX-cre, parvalbumin
INTRODUCTION
Among many brain regions, thalamic reticular nucleus (TRN) is known to be composed of exclusively GABAergic interneurons [9]. Thus, TRN neurons exert a potent inhibitory action through synaptically releasing GABA at their axon terminals [10,11,12]. On the other hand, tonic inhibition current has not been observed in TRN neurons [13]. This is consistent with the fact that TRN neurons do not express the subunits that mediate tonic inhibition current [14]. In contrast, TRN neurons receive synaptic input from cerebral cortex, in which cortical neurons possess δ subunitcontaining extra-synaptic GABAA receptors, which are critical for generating GABAA receptor-mediated tonic inhibition current [14,15]. According to a recent report, extracellular GABA in dentate gyrus can inhibit perforant pathway neurons projecting to this area via presynaptic GABAA receptor located in the axon terminals of the perforant fibers [16]. Therefore, GABAA receptors containing δ subunit in the axon terminals of the cortical neurons projecting to TRN might sense tonically released GABA in the TRN. Therefore, we raise a possibility that tonic GABA release might exist in TRN neurons and that it can be released through neuronal Best1.
In order to study the role of neuronal Best1 for tonic inhibition in TRN region, which contains heterogeneous interneuronal populations, we opted for gene-silencing technique for manipulating
MATERIALS AND METHODS
Mice were allowed free access to food and water, and were maintained under a 12-h light/dark cycle, with the light cycle beginning at 8:00 AM. Animals were cared for and handled in accordance with institutional guidelines of the Korea Institute of Science and Technology, Seoul, Korea. DLX5/6-cre [23] mice and C57BL6/J mice were used.
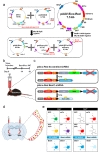
The general construction strategy for pSico-Red is described in Fig. 1a. For cre-dependent gene-silencing, U6-loxP-CMV-GFP-loxP-shRNA sequence from pSico lentiviral vector (addgene, UK) was amplified by PCR with MluI or BstB1-AflII-BglII-containing specific primers, and then it was cloned into pAAV-MCS vector (Stratagene, USA). To overcome disadvantage of pSico vector, cre-dependent removal of CMV-GFP sequence, additional EF1a promoter and mCherry sequences were sequentially added into MluI site of first cloned pAAV vector (pAAV-Sico) through the MluI, BstB1 and AflII restriction sites.
The Best1 nucleotides from 774 to 793(5'-tttgccaacttgtcaatgaa-3') was selected for target region of Best1 shRNA as previous report [5]. For adeno-associated-virus (AAV)-based shRNA expression, Best1 shRNA was synthesized as follows: 5'-tttgccaacttgtcaatgaat tcaagagatcattgacaagttggcaattttttc-3' and 5'-tcgagaaaaaatcgcatagcg tatgccgtttctcttgaaaacggcatacgctatgcgaa-3'. The annealed double-stranded oligo was inserted into HpaI-XhoI restriction enzyme sites of pAAV-Sico-Red vector and verified by sequencing. Scrambled shRNA-containing pAAV-Sico-Red construct was used as control. Using these viral vectors, AAV was packaged at KIST Virus Facility (http://virus.kist.re.kr).
Mice were anesthetized using 2% avertin. Mice were placed in a stereotaxic (Kopf instrument, USA) and the skull was exposed surgically. Bilateral craniotomies were performed using 0.5 mm diameter drill (SAESHIN, Korea) and the viruses were injected using a 33 gauge blunt needle (WPI, USA) or a glass micropipette attached to a 10 µl Hamilton micro syringe (Hamilton, Switzerland) filled with distilled water. A micro syringe pump (KD Scientific, USA) and its controller were used to maintain the injection speed of 0.25 µl/min. The needle was slowly lowered to the TRN (coordinate A.P.: 1.8, M.L.: 2.2, D.V.:3.7) and remained for 4 min after moving up and down for 3 times. Then, 2 µl of viruses (diluted by 1:3 ratio) are injected to the target site. After injection, the needle stayed for additional 4 minutes, before it was slowly withdrawn. Finally, mice's scalp was closed by 9 mm auto clip (MikRon Clay Adams, USA). Two weeks after virus injection, mice were sacrificed and brains were processed for immunohistochemistry (Fig. 1b).
Mice were euthanized by overdosing with 2% avertin and perfused transcardially with 30 ml of phosphate buffered saline (PBS), followed by 30 ml of 4% paraformaldehyde (Sigma-Aldrich, USA) dissolved in PBS. Brains were extracted from the skulls and incubated in 4% paraformaldehyde at 4℃ overnight. Brains were transferred to 30% sucrose solution. When brain sank to the bottom, brains were covered with optical cutting temperature (Scigen, Singapore) compound and frozen. Frozen brains were sliced at 30 µm coronal brain slices using a cryostat microtome (Thermo scientific, USA) and collected in PBS. Collected slices were rinsed in PBS 3 times, and incubated for 1hr 30 min with blocking solution (0.3% Triton-X, 2% normal goat serum, 2% normal donkey serum in 0.1 M PBS). Sections were incubated overnight in a mixture of the following primary antibodies with blocking solution at 4℃ on a shaker; rabbit anti-Parvalbumin antibody (1:500, Abcam, UK), rabbit anti-mCherry antibody (1:500, Abcam, UK), rabbit-anti-mouse bestrophin antibody, chicken anti-GFP antibody (1:500, Abcam, UK). After washing three times in PBS, sections were incubated with secondary antibodies; conjugated Alexa 405 donkey anti rabbit IgG (1:200, Jackson Immuno Research Laboratories, USA), conjugated Alexa 647 donkey anti Guinee pig IgG (1:200, Jackson Immuno Research Laboratories, USA), conjugated Alexa 594 donkey anti rabbit IgG (1:200, Jackson Immuno Research Laboratories, USA), conjugated Alexa 488 donkey anti chicken IgG (1:200, Jackson Immuno Research Laboratories, USA), for 1hr 30min, followed by three time rinses in PBS. And tissues were counterstained with DAPI (1:3000, Pierce, USA). They are mounted with fluorescent mounting medium (Dako, Denmark). A series of fluorescence images were obtained with Nikon A1 confocal microscopy.
Immunolabeled slices from 2 mice were analyzed for cell counting. Cells were visually inspected and manually counted. Neuronal cells are only counted. When counting cells, laser intensity was adjusted to exclude background negative signal for clearly obtaining viral infected cells as well as Best1 signal.
Student's t-test [24] was used to analyze the difference of the intensity of Best1 immunoreactivity. All values indicated are mean±SEM (standard error of mean) and the statistical significance is presented with n.s. (not significant) or asterisks: p values of <0.05(*), <0.01(**), and <0.001(***).
We developed pAAV-Sico-Red (pSico-Red) vector, which has another reporter gene,
To confirm if the pSico-Red system is working, we injected pSico-Red vector including Best1 or scrambled shRNA into TRN of DLX5/6-cre mice (Fig. 1b and c). We expected that in terms of the cells infected by pSico-Red Best1 shRNA, cre-negative cells would show both mCherry and GFP signal and cre-positive cells would show only mCherry signal due to excision of GFP, along with stop codon, hindering the Best1 shRNA expression (Fig. 1c, d, and e). Moreover, cre-positive cells would express decreased Best1 expression, compared to scrambled shRNA, because of the genesilencing effect of Best1 shRNA (Fig. 1e).
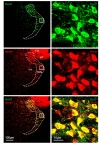
To test the possibility that Best1 is expressed in TRN region, we first performed immunohistochemistry with Best1 and NeuN antibodies. We, for the first time, found that Best1 is highly expressed in the neurons of TRN, indicated by NeuN-positive signals (Fig. 2). The percentage of TRN neurons expressing Best1 was almost 100%, whereas the neurons in the neighboring nuclei, stria terminalis and fimbria hippocampus, were mostly absent of Best1 immunoreactivity (Fig. 2). In marked contrast, we observed much lower level of Best1 in GFAP positive astrocytes than that in neurons (data is not shown). These results raise a possibility that, in addition to well-known astrocytic role, Best1 might have a neuronal function in the brain.
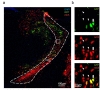
Based on the cloning strategy for pSico-Red, we predicted that cre-negative cells infected by AAV virus containing pSico-Red vector would express both mCherry and GFP signals, whereas crepositve cells would express only mCherry signal due to excision of GFP. To validate these possibilities, we analyzed TRN tissues of DLX5/6-cre mice, injected with AAV-pSico-Red virus using confocal microscope. DLX5/6-cre mouse was utilized due to its cell-type expression of cre in a subpopulation of GABAergic interneurons in hippocampus [25,26]. We observed that most of infected cells in TRN only expressed mCherry signal, implying that these cells are cre-expressing DLX5/6-positive neurons (Fig. 3a and b). However, we also found that a few TRN neurons expressed both mCherry and GFP signals, representing the crenegative neurons (Fig. 3a and b).
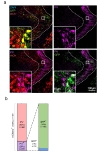
GABAergic interneurons can be subdivided into several classes, including parvalbumin positive neurons [27]. To determine the identity of DLX5/6-cre expressing neurons in TRN, we next performed immunohistochemistry with antibody against parvalbumin (PV). We found that most of the middle- and outerlayer TRN neurons were PV-positive, but not in inner-layer (Fig. 4). Among the mCherry positive neurons (or virus infected neurons) in TRN, about 20% of neurons were PV-positive, and remaining 80% was PV-negative (Fig. 4b). Of the PV-positive and mCherry-positive neurons, 95% turned out to be GFP-positive, which means that these neurons were DLX5/6-negative. In other words, DLX5/6 and PV label two non-overlapping populations of GABAergic interneurons in TRN (Fig. 4b). Taken together, we demonstrate that about 80% of neurons in TRN are DLX5/6-positive and PV-negative GABAergic interneurons, whereas remaining 20% neurons are DLX5/6-negative but PV-positive interneurons.
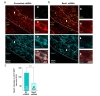
We have determined that TRN neurons express Best1 and that pSico-Red system identified a subpopulation of TRN neurons. To test the ability of pSico-Red to achieve cell-type-specific gene-silencing, we injected viral pSico-Red that contains either Best1-targeted shRNA or control scrambled shRNA into TRN of DLX5/6-cre mice (pSico-Red-Best1 or pSico-Red-SC). The specificity and efficiency of Best1-targeted shRNA have been reported previously in several reports [3,4,5,8]. Then the Best1 immunoreactivity was assessed in mCherry-positive but GFP-negative neurons (Fig. 5a and b). We found that the intensity of Best1 immunoreactivity was significantly decreased in Best1-shRNA group compared to that of scrambled shRNA group (Fig. 5c). These results indicate that pSico-Red is fully functional in cell-type-specific gene-silencing. Furthermore, these results confirm the specificity of Best1 antibody and imply that the immunoreactivity by Best1 antibody is genuinely labeling the Best1 protein.
In this study, we have developed advanced gene-silencing system, pSico-Red (Fig. 1). Using pSico-Red, we were able to identify the cells that express shRNA, which was not possible with the original pSico system. In addition, this system allowed us to determine the cell-type classification of cre-expressing neurons in TRN by assessing the distribution pattern of the two fluorescent reporters, mCherry and GFP (Fig. 2 and 3). In addition to these added features, we achieved cell-type-specific gene-silencing with targeted shRNA for Best1. Furthermore, one can combine pSico-Red with viruses carrying promoter specific cre genes in any animal model systems, where cre-expressing animal lines are not available. In this way, cell-type-specific gene-silencing can be achieved even without cre-expressing transgenic animals. In general, most of genes can be silenced in both region-specific and cell-type-specific way by using viruses for pSico-Red in combination with creexpressing transgenic mice.
The analysis of DLX5/6-cre mouse demonstrated that DLX5/6 defines a new subpopulation of GABAergic TRN interneurons that are non-overlapping and mutually exclusive with PV. Our results showed that there was only 1% of total TRN neurons and 5% of total PV-positive neurons that showed both DLX5/6 and PV expression (Fig. 4b). Previous study reported that about 80% of TRN neurons are somatostatin positive [28], and we found that about 20% of virus infected TRN neurons are PV positive (Fig. 4b). However, it is still unclear what proportion of these neurons are both somatostatin and PV positive. Our results imply that most of DLX5/6 positive neurons are somatostatin positive, given the fact that most DLX5/6 positive neurons are PV negative. Future work is needed to determine the exact extent of overlap between somatostatin and DLX5/6.
We have demonstrated for the first time that Best1 is highly expressed in almost all of TRN neurons, whereas Best1 is weakly expressed in TRN astrocytes. These results raise a possibility that Best1 might have a neuronal function in the brain. In TRN, we raise two potential functions of Best1: tonic GABA release and electrogenic role. Because TRN neurons are GABAergic neurons, Best1 is expected to play an important role in tonic release of GABA. Interestingly, TRN neurons themselves do not express high affinity, extrasynaptic GABAA receptors that mediate tonic inhibition current [13,14]. It follows that even if GABA is tonically released through Best1 expressed in TRN neurons, the autocrine action of tonic GABA would be minimal. We have recently reported that tonic GABA in dentate gyrus can inhibit synaptic release of glutamate from perforant fibers via presynaptic GABAA and GABAB receptors located in the axon terminals of the perforant fibers [16]. Therefore, we expect that tonic GABA would exert a potent effect on GABAA and GABAB that are expressed at presynaptic terminals that terminate onto TRN neurons. Thus, tonic GABA released from TRN neurons would have strong inhibitory role on presynaptic release of various transmitters including glutamate, in particular, released onto TRN neurons. For example, major projection of cortical neurons to TRN neurons is glutamatergic and this projection could be heavily controlled by tonic GABA release from TRN neurons. Future studies are needed to test these possibilities.
In addition to the possible role of Best1 in tonic GABA release, Best1 could have an electrogenic role in TRN neurons. TRN neurons express high level of voltage-gated Ca2+ channels, including Cav3.2 and Cav3.3, which play a critical role in firing pattern and in absence seizure [29]. Because Best1 is a Ca2+-activated chloride channel, Best1 can be activated upon activation of voltage-gated Ca2+ channels. In addition to Ca2+-activated potassium channels, such as SK1 and SK2, Best1 could control neuronal excitability, action potential shape and time course, and rhythmic burst discharges of TRN neurons. Future studies are needed to test these exciting possibilities.
In summary, the present study provides technical advancement in gene-silencing molecular tool by developing pSico-Red, and first evidence for neuronal expression of Best1. In addition, we have identified a novel subpopulation of DLX5/6 expressing, PV-negative GABAergic interneurons in TRN. The function of neuronal Best1 in TRN remains to be explored, but the results and pSico-Red described here should provide an opportunity to address many interesting questions regarding the role of neuronal Best1 in brain function.
- Hartzell HC, Qu Z, Yu K, Xiao Q, Chien LT. Molecular physiology of bestrophins: multifunctional membrane proteins linked to best disease and other retinopathies. Physiol Rev 2008;88:639-672.
- Petrukhin K, Koisti MJ, Bakall B, Li W, Xie G, Marknell T, Sandgren O, Forsman K, Holmgren G, Andreasson S, Vujic M, Bergen AA, McGarty-Dugan V, Figueroa D, Austin CP, Metzker ML, Caskey CT, Wadelius C. Identification of the gene responsible for Best macular dystrophy. Nat Genet 1998;19:241-247.
- Jo S, Yarishkin O, Hwang YJ, Chun YE, Park M, Woo DH, Bae JY, Kim T, Lee J, Chun H, Park HJ, Lee Y, Hong J, Kim HY, Oh SJ, Park SJ, Lee H, Yoon BE, Kim Y, Jeong Y, Shim I, Bae YC, Cho J, Kowall NW, Ryu H, Hwang E, Kim D, Lee CJ. GABA from reactive astrocytes impairs memory in mouse models of Alzheimer's disease. Nat Med 2014;20:886-896.
- Lee S, Yoon BE, Berglund K, Oh SJ, Park H, Shin HS, Augustine GJ, Lee CJ. Channel-mediated tonic GABA release from glia. Science 2010;330:790-796.
- Park H, Oh SJ, Han KS, Woo DH, Park H, Mannaioni G, Traynelis SF, Lee CJ. Bestrophin-1 encodes for the Ca2+-activated anion channel in hippocampal astrocytes. J Neurosci 2009;29:13063-13073.
- Yoon BE, Jo S, Woo J, Lee JH, Kim T, Kim D, Lee CJ. The amount of astrocytic GABA positively correlates with the degree of tonic inhibition in hippocampal CA1 and cerebellum. Mol Brain 2011;4:42.
- Woo DH, Han KS, Shim JW, Yoon BE, Kim E, Bae JY, Oh SJ, Hwang EM, Marmorstein AD, Bae YC, Park JY, Lee CJ. TREK-1 and Best1 channels mediate fast and slow glutamate release in astrocytes upon GPCR activation. Cell 2012;151:25-40.
- Park H, Han KS, Oh SJ, Jo S, Woo J, Yoon BE, Lee CJ. High glutamate permeability and distal localization of Best1 channel in CA1 hippocampal astrocyte. Mol Brain 2013;6:54.
- Yen CT, Conley M, Hendry SH, Jones EG. The morphology of physiologically identified GABAergic neurons in the somatic sensory part of the thalamic reticular nucleus in the cat. J Neurosci 1985;5:2254-2268.
- Cox CL, Huguenard JR, Prince DA. Heterogeneous axonal arborizations of rat thalamic reticular neurons in the ventrobasal nucleus. J Comp Neurol 1996;366:416-430.
- De Biasi S, Frassoni C, Spreafico R. The intrinsic organization of the ventroposterolateral nucleus and related reticular thalamic nucleus of the rat: a double-labeling ultrastructural investigation with gamma-aminobutyric acid immunogold staining and lectin-conjugated horseradish peroxidase. Somatosens Res 1988;5:187-203.
- Pinault D, Bourassa J, Deschênes M. The axonal arborization of single thalamic reticular neurons in the somatosensory thalamus of the rat. Eur J Neurosci 1995;7:31-40.
- Belelli D, Peden DR, Rosahl TW, Wafford KA, Lambert JJ. Extrasynaptic GABAA receptors of thalamocortical neurons: a molecular target for hypnotics. J Neurosci 2005;25:11513-11520.
- Cope DW, Hughes SW, Crunelli V. GABAA receptor-mediated tonic inhibition in thalamic neurons. J Neurosci 2005;25:11553-11563.
- Sherman SM, Guillery RW. Functional organization of thalamocortical relays. J Neurophysiol 1996;76:1367-1395.
- Yarishkin O, Lee J, Jo S, Hwang EM, Lee CJ. Disinhibitory action of astrocytic GABA at the perforant path to dentate gyrus granule neuron synapse reverses to inhibitory in Alzheimer's disease model. Exp Neurobiol 2015;24:211-218.
- Utomo AR, Nikitin AY, Lee WH. Temporal, spatial, and cell type-specific control of Cre-mediated DNA recombination in transgenic mice. Nat Biotechnol 1999;17:1091-1096.
- Saurabh S, Vidyarthi AS, Prasad D. RNA interference: concept to reality in crop improvement. Planta 2014;239:543-564.
- Ventura A, Meissner A, Dillon CP, McManus M, Sharp PA, Van Parijs L, Jaenisch R, Jacks T. Cre-lox-regulated conditional RNA interference from transgenes. Proc Natl Acad Sci U S A 2004;101:10380-10385.
- Lee Y, Morrison BM, Li Y, Lengacher S, Farah MH, Hoffman PN, Liu Y, Tsingalia A, Jin L, Zhang PW, Pellerin L, Magistretti PJ, Rothstein JD. Oligodendroglia metabolically support axons and contribute to neurodegeneration. Nature 2012;487:443-448.
- Young SZ, Taylor MM, Wu S, Ikeda-Matsuo Y, Kubera C, Bordey A. NKCC1 knockdown decreases neuron production through GABA(A)-regulated neural progenitor proliferation and delays dendrite development. J Neurosci 2012;32:13630-13638.
- Heitz F, Johansson T, Baumgärtel K, Gecaj R, Pelczar P, Mansuy IM. Heritable and inducible gene knockdown in astrocytes or neurons in vivo by a combined lentiviral and RNAi approach. Front Cell Neurosci 2014;8:62.
- Stenman J, Toresson H, Campbell K. Identification of two distinct progenitor populations in the lateral ganglionic eminence: implications for striatal and olfactory bulb neurogenesis. J Neurosci 2003;23:167-174.
- Sokal RR, Rohlf FJ. Taxonomic congruence in the Leptopodomorpha re-examined. Syst Zool 1981;30:309-325.
- Monory K, Massa F, Egertová M, Eder M, Blaudzun H, Westenbroek R, Kelsch W, Jacob W, Marsch R, Ekker M, Long J, Rubenstein JL, Goebbels S, Nave KA, During M, Klugmann M, Wölfel B, Dodt HU, Zieglgänsberger W, Wotjak CT, Mackie K, Elphick MR, Marsicano G, Lutz B. The endocannabinoid system controls key epileptogenic circuits in the hippocampus. Neuron 2006;51:455-466.
- Dine J, Kühne C, Deussing JM, Eder M. Optogenetic evocation of field inhibitory postsynaptic potentials in hippocampal slices: a simple and reliable approach for studying pharmacological effects on GABAA and GABAB receptor-mediated neurotransmission. Front Cell Neurosci 2014;8:2.
- Cowan RL, Wilson CJ, Emson PC, Heizmann CW. Parvalbumin-containing GABAergic interneurons in the rat neostriatum. J Comp Neurol 1990;302:197-205.
- Ahrens S, Jaramillo S, Yu K, Ghosh S, Hwang GR, Paik R, Lai C, He M, Huang ZJ, Li B. ErbB4 regulation of a thalamic reticular nucleus circuit for sensory selection. Nat Neurosci 2015;18:104-111.
- Lee SE, Lee J, Latchoumane C, Lee B, Oh SJ, Saud ZA, Park C, Sun N, Cheong E, Chen CC, Choi EJ, Lee CJ, Shin HS. Rebound burst firing in the reticular thalamus is not essential for pharmacological absence seizures in mice. Proc Natl Acad Sci U S A 2014;111:11828-11833.



