Articles
Article Tools
Stats or Metrics
Article
Original Article
Exp Neurobiol 2016; 25(5): 269-276
Published online October 31, 2016
https://doi.org/10.5607/en.2016.25.5.269
© The Korean Society for Brain and Neural Sciences
LRRK2 Inhibits FAK Activity by Promoting FERM-mediated Autoinhibition of FAK and Recruiting the Tyrosine Phosphatase, SHP-2
Insup Choi1,2,3,4, Ji-won Byun1, Sang Myun Park1,2,4, Ilo Jou1,2,4 and Eun-Hye Joe1,2,3,4*
1Department of Biomedical Sciences, Neuroscience Graduate Program, 2Department of Pharmacology, 3Department of Brain Science, 4Chronic Inflammatory Disease Research Center, Ajou University School of Medicine, Suwon 16499, Korea
Correspondence to: *To whom correspondence should be addressed.
TEL: 82-31-219-5062, FAX: 82-31-219-5069
e-mail: ehjoe@ajou.ac.kr
Mutation of leucine-rich repeat kinase 2 (LRRK2) causes an autosomal dominant and late-onset familial Parkinson's disease (PD). Recently, we reported that LRRK2 directly binds to and phosphorylates the threonine 474 (T474)-containing Thr-X-Arg(Lys) (TXR) motif of focal adhesion kinase (FAK), thereby inhibiting the phosphorylation of FAK at tyrosine (Y) 397 residue (pY397-FAK), which is a marker of its activation. Mechanistically, however, it remained unclear how T474-FAK phosphorylation suppressed FAK activation. Here, we report that T474-FAK phosphorylation could inhibit FAK activation via at least two different mechanisms. First, T474 phosphorylation appears to induce a conformational change of FAK, enabling its N-terminal FERM domain to autoinhibit Y397 phosphorylation. This is supported by the observation that the levels of pY397-FAK were increased by deletion of the FERM domain and/or mutation of the FERM domain to prevent its interaction with the kinase domain of FAK. Second, pT474-FAK appears to recruit SHP-2, which is a phosphatase responsible for dephosphorylating pY397-FAK. We found that mutation of T474 into glutamate (T474E-FAK) to mimic phosphorylation induced more strong interaction with SHP-2 than WT-FAK, and that pharmacological inhibition of SHP-2 with NSC-87877 rescued the level of pY397 in HEK293T cells. These results collectively show that LRRK2 suppresses FAK activation through diverse mechanisms that include the promotion of autoinhibition and/or the recruitment of phosphatases, such as SHP-2.
Keywords: Parkinson’s disease, LRRK2, FAK, phosphatase, SHP-2
Focal adhesion kinase (FAK) is a non-receptor kinase that controls the migration, proliferation, and survival of cells [13,14,15]. It consists of an N-terminal FERM domain, a kinase domain, and a C-terminal focal adhesion-targeting (FAT) domain [16,17]. During cell migration, FAK is activated and recruited to the focal adhesion sites where lamellipodia are produced; this activates downstream signaling molecules that regulate the reorganization of cytoskeletal proteins, including the polymerization of actin [15]. FAK can be activated in response to cell-migration-promoting stimuli, such as the interaction between the extracellular matrix (ECM) and integrin [18], the activation of growth factor receptors or G protein-coupled receptors [19], and mechanical stress [20]. Upon activation of FAK demonstrated by autophosphorylation of Y397 (pY397), downstream signaling is activated for proper cell migration [15,21].
We recently showed that G2019S-LRRK2 strongly inhibits FAK and attenuates microglial motility [9]. Our results revealed that microglia derived from G2019S-LRRK2 transgenic mice (TG-microglia) exhibited impaired FAK activation (decreased levels of pY397) when treated with ADP, which is a microglial activator that increases motility. TG-microglia produced unstable lamellipodia and exhibited reduce motility compared with wild-type (WT)-microglia. Moreover, we found that LRRK2 suppresses FAK activation by directly phosphorylating the Thr residue(s) in the Thr-X-Arg (TXR) motif(s) of FAK, which include Thr 474 (T474). In the present study, we further examined how T474-FAK phosphorylation prevents the activation of FAK. Our novel results suggest that T474 phosphorylation may promote the FERM-mediated autoinhibition of FAK and/or trigger the recruitment of SHP-2, which dephosphorylates pY397-FAK. Thus, LRRK2 appears to regulate FAK activity through diverse mechanisms.
MATERIALS AND METHODS
The HEK293T cell line was acquired from ATCC (Seoul, Korea), and maintained in DMEM supplemented with 10% (v/v) FBS and penicillin/streptomycin (50 U/mL).
FLAG-FAK was prepared by inserting the human FAK gene into the p3xFLAG-CMV-7.1 vector (Sigma, St Louis, MO, USA) using AccuPrime Pfx DNA Polymerase (Invitrogen, Carlsbad, CA, USA) and an infusion cloning kit (Clontech, Palo Alto, CA, USA). Mutations were introduced into FLAG-FAK using a QuikChange Lightning Site-Directed Mutagenesis Kit (Agilent Technologies, Palo Alto, CA, USA). The FERM domain deletion mutant (Δ35~362) was prepared using AccuPrime Pfx DNA Polymerase. Plasmids encoding WT-SHP-2 were kindly provided by Prof. Young Ho Suh (Seoul National University College of Medicine, Seoul, Korea). The primers used for mutagenesis are listed in Table 1.
HEK293T cells were transfected with DNA plasmids using the jetPEI transfection reagent (Polyplus-Transfection, San Diego, CA, USA) as described by the manufacturer. Briefly, cells were exposed to DNA plasmids and the jetPEI mixture for 4 hours, and then the media were replaced with fresh DMEM containing 10% FBS. Two days later, transfected cells were used for experiments.
Cells were lysed on ice in RIPA buffer (50 mM Tris-HCl pH 7.4, 1% NP-40, 1 mM NaF, 0.25% Na-deoxycholate, 1 mM Na3VO4, and 150 mM NaCl) containing a protease/phosphatase inhibitor cocktail (GenDEPOT, Barker, TX, USA). Lysates were centrifuged, and the proteins in the supernatant were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and blotted to nitrocellulose membranes (Protran; Schleicher & Schuell, Dassel, Germany). Membranes were incubated with antibodies specific for FAK (1:1000, Santa Cruz, Dallas, TX, USA), pY397-FAK (1:1000, Cell Signaling Technology, Danvers, MA, USA), FLAG (1:2000, Sigma), SHP1 (1:1000, BD Bioscience, NJ, USA), SHP2 (1:1000, BD Bioscience), PTPD1 (1:1000, Thermo Scientific), and PTP-PEST (1:1000, Cell Signaling Technology). The membranes were washed with Tris-buffered saline containing 0.1% Tween 20 (TBST) and incubated with secondary antibodies, and the results were visualized using an enhanced chemiluminescence system (Daeil Lab Inc., Seoul, Korea).
WT- and T474E-FAK-encoding constructs were transfected to HEK293T cells. After 2 days, the cells were lysed with an immunoprecipitation buffer (1% Triton X-100, 150 mM NaCl, 10 mM NaH2PO4, 15 mM Na2HPO4, 50 mM NaF, 1 mM EDTA, and 1 mM Na3VO4). Cell lysates (500 µg) were incubated with primary antibodies against FLAG (1 µg, F1804; Sigma) and then with Protein G agarose beads (20 µl per reaction; Millipore, Billerica, MA, USA). The protein-antibody-bead complexes were treated with 100 µM FLAG peptide (Sigma) in 50 mM Tris-HCl pH 7.4 to release FLAG-FAK proteins from the agarose beads. The antibody-protein G agarose complexes were removed by centrifugation, and equal volumes of supernatants were subjected to Western blotting.
FLAG-tagged WT and mutant FAK proteins were expressed in HEK293T cells, immunoprecipitated, and isolated from the agarose beads as described above. The released FLAG-FAK proteins were incubated in kinase buffer S (50 mM Tris-HCl, pH 8.5, 10 mM MgCl2, 0.01% Brij-35, and 1 mM EGTA; Invitrogen) containing a protease/phosphatase inhibitor cocktail (GenDEPOT) and 10 µM ATP and/or 1 µCi/mL 32P-ATP (Perkin Elmer-Cetus, Norwalk, CT, USA).
The band intensities of the Western blots and Coomassie blue-stained gels were quantified using the Image J software (NIH, Bethesda, MD, USA). The statistical significance of differences was determined by one-way analysis of variance (ANOVA) followed by the Newman-Keuls post hoc test, as applied using the Graph Pad Prism 5 software package (GraphPad Software, San Diego, CA, USA).
We previously reported that FAK has six TXR consensus motifs that may be phosphorylated by LRRK2, and that the among phosphorylation-mimicking mutation of Thr (T) to Glu (E) (T→E), only T474 mutation attenuated the levels of pY397 in HEK293T cells [9]. Since the activation of FAK in these cells could be controlled by diverse mechanisms, we used
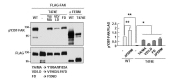
Next, we sought to determine how the phosphorylation of T474 suppresses FAK activity. A previous study showed that the FERM domain of FAK physically covers Y397, thereby blocking its autophosphorylation [22]. Conversely, point mutations of V180A/M183A, V196D/L197D, and/or F596D were shown to prevent the interaction between the FERM and kinase domains and increase Y397 autophosphorylation [23]. Here, we analyzed whether T474 phosphorylation decreased FAK activity through the FERM domain. Interestingly, all three point mutations, V180A/M183A, V196D/L197D, and F596D, rescued Y397 phosphorylation in T474E-FAK-overexpressing HEK293 T cells (Fig. 2). Furthermore, deletion of the FERM domain (ΔFERM) increased the pY397 levels for both WT-FAK and T474E-FAK (Fig. 2). These findings suggest that T474 phosphorylation induces FERM-mediated autoinhibition, thereby decreasing FAK activity.
FAK activity is known to be regulated by phosphatases that dephosphorylate pY397, such as protein tyrosine phosphatase (PTP)-PEST [24,25], PTPD1 [26], SHP-1 [27], and SHP-2 [28,29]. Accordingly, we treated T474E-FAK-overexpressing HEK293T cells with the broad-spectrum tyrosine phosphatase inhibitor, pervanadate [30], and examined whether phosphatases are involved in the T474-phosphorylation-induced decrease in pY397-FAK. Interestingly, we found that the level of pY397-FAK was rescued by pervanadate treatment (Fig. 3A). Previously, we reported that overexpression of G2019S-LRRK2 in HEK293T cells weakly increased pY397-FAK levels in response to ADP compared with overexpression of WT- or D1994A (a kinase dead mutant)-LRRK2 [9]. Here, we found that pervanadate rescued pY397-FAK levels in G2019S-LRRK2-overexpressing cells (Fig. 3B). These results suggest that tyrosine phosphatases contribute to the decreases in pY397 levels induced by T474 phosphorylation mimicked by overexpression of T474E-FAK or G2019S-LRRK2.
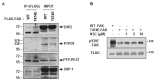
Next, we examined which tyrosine phosphatase(s) is/are responsible for dephosphorylating pY397-FAK. Our immunoprecipitation experiments revealed that T474E-FAK associated more strongly with SHP-2 than did WT-FAK (Fig. 4A). In contrast, PTPD1 and PTP-PEST both failed to bind WT- or T474E-FAK, while SHP-1 appeared to bind WT- and T474E-FAK with similar intensities (Fig. 4A). Our subsequent experiments revealed that the SHP-1/2 inhibitor, NSC-87877 (NSC), increased the level of pY397 in T474E-FAK-overexpressing HEK293T cells (Fig. 4B).
Taken together, our present findings suggest that the LRRK2-mediated phosphorylation of FAK at T474 suppresses FAK activity by inducing its autoinhibition and/or recruiting SHP-2 to dephosphorylate pY397.
FAK, which is known to regulate diverse and fundamental biological phenomena (e.g., cell survival, proliferation, and migration), is highly expressed in the brain, neurons, and glial cells, such as microglia and astrocytes [9,31,32]. Previous studies showed that conditional knock-out of FAK in neurons during postnatal brain development increased the number of axon terminals and synapses by increasing branch formation [33], while that in cultured hippocampal neurons triggered defects in spine formation [34]. FAK has been shown to mediate neurotrophin-induced neurite outgrowth in hippocampal neurons, and is known to regulate miniature excitatory postsynaptic currents (mEPSCs), long-term potentiation (LTP), and hippocampus-mediated spatial learning and memory [35]. In addition, FAK regulates the myelination of oligodendrocytes: conditional deletion of FAK in oligodendrocytes decreased the presence of myelinated fibers during development, but myelination became normal after birth, suggesting that FAK is involved in the initiation of myelination [36]. FAK inhibitors reportedly cause defects in microglial motility [9], which is important for the ability of these cells to scan the brain environment [37,38,39,40]. The PD-associated pathogenic LRRK2-G2019S mutant may affect neurons and non-neuronal cells in many ways through FAK. We previously showed that G2019S-LRRK2 directly phosphorylates FAK and decreases microglial motility [9], and other groups have reported that G2019S-LRRK2 suppresses neurite outgrowth by regulating ezrin/radixin/moesin (ERM) [6,41], Rac1 [42], Rab5 [43], and actin-related molecules [7]. FAK functions as a platform for downstream cascades that regulate actin/microtubule polymerization/depolymerization [15]. FAK also mediates the translocation of Rac1 to adhesion sites to promote lamellipodia extension and cell spreading [44], and interacts with actin-related protein complex (Arp2/3) to induce actin nucleation as an initial step for actin polymerization. Therefore, it has been suggested that G2019S-LRRK2 affects the functions and/or properties of neurons or non-neuronal cells by regulating FAK.
The activity of FAK is regulated by its phosphorylation state. In response to stimuli that induce cell motility, FAK undergoes autophosphorylation at Y397, which triggers its Src-mediated phosphorylation at Y576/577, Y861, Y863, and Y925 [45,46,47,48]. Previous studies have shown that: FAK Y576/Y577 phosphorylation is associated with maximum FAK activity [46]; the VEGF-mediated phosphorylation of Y861 promotes the formation of the FAK-integrin αvβ5 complex and facilitates migration in endothelial cells [49,50]; Y861 phosphorylation enhances FAK Y397 autophosphorylation [45]; Y925 positively regulates lamellipodia formation and cell migration by promoting focal adhesion disassembly [47]; and the Cdk5-mediated phosphorylation of S732 is necessary for neuronal migration [51]. We previously added to this body of knowledge by showing that the LRRK2-mediated phosphorylation of T474 negatively regulates FAK activity by reducing Y397 phosphorylation [9]. Since LRRK2 is a Ser/Thr kinase, we speculated that one or more complicated mechanism(s) could be involved in the LRRK2-mediated inhibition of FAK Y397 phosphorylation. Here, we report for the first time that FERM domain point mutations and deletions previously shown to uncover the kinase domain [23] could rescue Y397 phosphorylation in T474E-FAK-overexpressing cells. Furthermore, the pY397 level of T474E-FAK was significantly reduced by SHP-2. Collectively, these results suggest a new mechanism for the regulation of FAK activation, wherein FAK T474 phosphorylation inhibits the phosphorylation of Y397 through FERM-mediated autoinhibition and/or SHP-2-mediated Y397 dephosphorylation.


| Mutation | Primer sequence(s) |
|---|---|
| FAK | |
| T474A | 5’-CTTCAAGAAGCCTTAGCGATGCGTCAGTTTGACCATCCTC-3’ |
| T219E | 5’-GGATTCTGTCAAGGCCAAAGAGCTAAGAAAACTGATCC-3’ |
| T227E | 5’-CTAAGAAAACTGATCCAACAAGAGTTTAGACAATTTGCCAACC-3’ |
| T284E | 5’-AATCAGTTACCTAGAGGACAAGGGCTGCAATCCC-3’ |
| T455E | 5’-CGGTTGCAATTAAAGAGTGTAAAAACTGTACTTCGGACAGCG-3’ |
| T474E | 5’-CTTCAAGAAGCCTTAGAGATGCGTCAGTTTGACCATCCTC-3’ |
| T979E | 5’-TACCAGCCAGCGAGCACCGAGAGATTGAGATGGC-3’ |
| Y180A/M183A | 5’-CGGCGATCAGCCTGGGAGGCGCGGGGCAATGC-3’ |
| V196D/L197D | 5’-GAAAAGAAGTCTAACTATGAAGATGATGAAAAAGATGTTGGT TTAAAGCG-3’ |
| F596D | 5’-TGGCTCCAGAGTCAATCAATGATCGACGTT TTACCTCAGC-3’ |
| FERM domain deletion (Δ35~362) | 5’-CTCCATTGCACCAGGAGAACGTTCC-3’ and 5’-CAGAAAGAAGGTGAACGGGCTTTGCC-3' |
- Mata IF, Wedemeyer WJ, Farrer MJ, Taylor JP, Gallo KA. LRRK2 in Parkinson's disease: protein domains and functional insights. Trends Neurosci 2006;29:286-293.
- Paisán-Ruiz C, Washecka N, Nath P, Singleton AB, Corder EH. Parkinson's disease and low frequency alleles found together throughout LRRK2. Ann Hum Genet 2009;73:391-403.
- Lee BD, Dawson VL, Dawson TM. Leucine-rich repeat kinase 2 (LRRK2) as a potential therapeutic target in Parkinson's disease. Trends Pharmacol Sci 2012;33:365-373.
- Lesage S, Dürr A, Tazir M, Lohmann E, Leutenegger AL, Janin S, Pollak P, Brice A, French Parkinson's Disease Genetics Study Group. LRRK2 G2019S as a cause of Parkinson's disease in North African Arabs. N Engl J Med 2006;354:422-423.
- Ozelius LJ, Senthil G, Saunders-Pullman R, Ohmann E, Deligtisch A, Tagliati M, Hunt AL, Klein C, Henick B, Hailpern SM, Lipton RB, Soto-Valencia J, Risch N, Bressman SB. LRRK2 G2019S as a cause of Parkinson's disease in Ashkenazi Jews. N Engl J Med 2006;354:424-425.
- Parisiadou L, Xie C, Cho HJ, Lin X, Gu XL, Long CX, Lobbestael E, Baekelandt V, Taymans JM, Sun L, Cai H. Phosphorylation of ezrin/radixin/moesin proteins by LRRK2 promotes the rearrangement of actin cytoskeleton in neuronal morphogenesis. J Neurosci 2009;29:13971-13980.
- Meixner A, Boldt K, Van Troys M, Askenazi M, Gloeckner CJ, Bauer M, Marto JA, Ampe C, Kinkl N, Ueffing M. A QUICK screen for Lrrk2 interaction partners--leucine-rich repeat kinase 2 is involved in actin cytoskeleton dynamics. Mol Cell Proteomics 2011;10:M110.001172.
- Martin I, Kim JW, Lee BD, Kang HC, Xu JC, Jia H, Stankowski J, Kim MS, Zhong J, Kumar M, Andrabi SA, Xiong Y, Dickson DW, Wszolek ZK, Pandey A, Dawson TM, Dawson VL. Ribosomal protein s15 phosphorylation mediates LRRK2 neurodegeneration in Parkinson's disease. Cell 2014;157:472-485.
- Choi I, Kim B, Byun JW, Baik SH, Huh YH, Kim JH, Mook-Jung I, Song WK, Shin JH, Seo H, Suh YH, Jou I, Park SM, Kang HC, Joe EH. LRRK2 G2019S mutation attenuates microglial motility by inhibiting focal adhesion kinase. Nat Commun 2015;6:8255.
- Cirnaru MD, Marte A, Belluzzi E, Russo I, Gabrielli M, Longo F, Arcuri L, Murru L, Bubacco L, Matteoli M, Fedele E, Sala C, Passafaro M, Morari M, Greggio E, Onofri F, Piccoli G. LRRK2 kinase activity regulates synaptic vesicle trafficking and neurotransmitter release through modulation of LRRK2 macro-molecular complex. Front Mol Neurosci 2014;7:49.
- Imai Y, Gehrke S, Wang HQ, Takahashi R, Hasegawa K, Oota E, Lu B. Phosphorylation of 4E-BP by LRRK2 affects the maintenance of dopaminergic neurons in Drosophila. EMBO J 2008;27:2432-2443.
- Liu GH, Qu J, Suzuki K, Nivet E, Li M, Montserrat N, Yi F, Xu X, Ruiz S, Zhang W, Wagner U, Kim A, Ren B, Li Y, Goebl A, Kim J, Soligalla RD, Dubova I, Thompson J, Yates J, Esteban CR, Sancho-Martinez I, Izpisua Belmonte JC. Progressive degeneration of human neural stem cells caused by pathogenic LRRK2. Nature 2012;491:603-607.
- Lim ST, Chen XL, Lim Y, Hanson DA, Vo TT, Howerton K, Larocque N, Fisher SJ, Schlaepfer DD, Ilic D. Nuclear FAK promotes cell proliferation and survival through FERM-enhanced p53 degradation. Mol Cell 2008;29:9-22.
- Yamamoto D, Sonoda Y, Hasegawa M, Funakoshi-Tago M, Aizu-Yokota E, Kasahara T. FAK overexpression upregulates cyclin D3 and enhances cell proliferation via the PKC and PI3-kinase-Akt pathways. Cell Signal 2003;15:575-583.
- Mitra SK, Hanson DA, Schlaepfer DD. Focal adhesion kinase: in command and control of cell motility. Nat Rev Mol Cell Biol 2005;6:56-68.
- Dunty JM, Gabarra-Niecko V, King ML, Ceccarelli DF, Eck MJ, Schaller MD. FERM domain interaction promotes FAK signaling. Mol Cell Biol 2004;24:5353-5368.
- Zheng C, Xing Z, Bian ZC, Guo C, Akbay A, Warner L, Guan JL. Differential regulation of Pyk2 and focal adhesion kinase (FAK). The C-terminal domain of FAK confers response to cell adhesion. J Biol Chem 1998;273:2384-2389.
- Guan JL, Trevithick JE, Hynes RO. Fibronectin/integrin interaction induces tyrosine phosphorylation of a 120-kDa protein. Cell Regul 1991;2:951-964.
- Sieg DJ, Hauck CR, Ilic D, Klingbeil CK, Schaefer E, Damsky CH, Schlaepfer DD. FAK integrates growth-factor and integrin signals to promote cell migration. Nat Cell Biol 2000;2:249-256.
- Shyy JY, Chien S. Role of integrins in cellular responses to mechanical stress and adhesion. Curr Opin Cell Biol 1997;9:707-713.
- Parsons JT, Martin KH, Slack JK, Taylor JM, Weed SA. Focal adhesion kinase: a regulator of focal adhesion dynamics and cell movement. Oncogene 2000;19:5606-5613.
- Jácamo RO, Rozengurt E. A truncated FAK lacking the FERM domain displays high catalytic activity but retains responsiveness to adhesion-mediated signals. Biochem Biophys Res Commun 2005;334:1299-1304.
- Lietha D, Cai X, Ceccarelli DF, Li Y, Schaller MD, Eck MJ. Structural basis for the autoinhibition of focal adhesion kinase. Cell 2007;129:1177-1187.
- Zheng Y, Yang W, Xia Y, Hawke D, Liu DX, Lu Z. Ras-induced and extracellular signal-regulated kinase 1 and 2 phosphorylation-dependent isomerization of protein tyrosine phosphatase (PTP)-PEST by PIN1 promotes FAK dephosphorylation by PTP-PEST. Mol Cell Biol 2011;31:4258-4269.
- Zheng Y, Xia Y, Hawke D, Halle M, Tremblay ML, Gao X, Zhou XZ, Aldape K, Cobb MH, Xie K, He J, Lu Z. FAK phosphorylation by ERK primes ras-induced tyrosine dephosphorylation of FAK mediated by PIN1 and PTP-PEST. Mol Cell 2009;35:11-25.
- Carlucci A, Gedressi C, Lignitto L, Nezi L, Villa-Moruzzi E, Avvedimento EV, Gottesman M, Garbi C, Feliciello A. Protein-tyrosine phosphatase PTPD1 regulates focal adhesion kinase autophosphorylation and cell migration. J Biol Chem 2008;283:10919-10929.
- Lin SY, Raval S, Zhang Z, Deverill M, Siminovitch KA, Branch DR, Haimovich B. The protein-tyrosine phosphatase SHP-1 regulates the phosphorylation of alpha-actinin. J Biol Chem 2004;279:25755-25764.
- Wang FM, Liu HQ, Liu SR, Tang SP, Yang L, Feng GS. SHP-2 promoting migration and metastasis of MCF-7 with loss of E-cadherin, dephosphorylation of FAK and secretion of MMP-9 induced by IL-1beta in vivo and in vitro. Breast Cancer Res Treat 2005;89:5-14.
- Hartman ZR, Schaller MD, Agazie YM. The tyrosine phosphatase SHP2 regulates focal adhesion kinase to promote EGF-induced lamellipodia persistence and cell migration. Mol Cancer Res 2013;11:651-664.
- Huyer G, Liu S, Kelly J, Moffat J, Payette P, Kennedy B, Tsaprailis G, Gresser MJ, Ramachandran C. Mechanism of inhibition of protein-tyrosine phosphatases by vanadate and pervanadate. J Biol Chem 1997;272:843-851.
- Grant SG, Karl KA, Kiebler MA, Kandel ER. Focal adhesion kinase in the brain: novel subcellular localization and specific regulation by Fyn tyrosine kinase in mutant mice. Genes Dev 1995;9:1909-1921.
- Krady JK, Basu A, Levison SW, Milner RJ. Differential expression of protein tyrosine kinase genes during microglial activation. Glia 2002;40:11-24.
- Rico B, Beggs HE, Schahin-Reed D, Kimes N, Schmidt A, Reichardt LF. Control of axonal branching and synapse formation by focal adhesion kinase. Nat Neurosci 2004;7:1059-1069.
- Shi Y, Pontrello CG, DeFea KA, Reichardt LF, Ethell IM. Focal adhesion kinase acts downstream of EphB receptors to maintain mature dendritic spines by regulating cofilin activity. J Neurosci 2009;29:8129-8142.
- Monje FJ, Kim EJ, Pollak DD, Cabatic M, Li L, Baston A, Lubec G. Focal adhesion kinase regulates neuronal growth, synaptic plasticity and hippocampus-dependent spatial learning and memory. Neurosignals 2012;20:1-14.
- Forrest AD, Beggs HE, Reichardt LF, Dupree JL, Colello RJ, Fuss B. Focal adhesion kinase (FAK): a regulator of CNS myelination. J Neurosci Res 2009;87:3456-3464.
- Nimmerjahn A, Kirchhoff F, Helmchen F. Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science 2005;308:1314-1318.
- Davalos D, Grutzendler J, Yang G, Kim JV, Zuo Y, Jung S, Littman DR, Dustin ML, Gan WB. ATP mediates rapid microglial response to local brain injury in vivo. Nat Neurosci 2005;8:752-758.
- Hines DJ, Hines RM, Mulligan SJ, Macvicar BA. Microglia processes block the spread of damage in the brain and require functional chloride channels. Glia 2009;57:1610-1618.
- Jeong HK, Ji KM, Kim B, Kim J, Jou I, Joe EH. Inflammatory responses are not sufficient to cause delayed neuronal death in ATP-induced acute brain injury. PLoS One 2010;5:e13756.
- Jaleel M, Nichols RJ, Deak M, Campbell DG, Gillardon F, Knebel A, Alessi DR. LRRK2 phosphorylates moesin at threonine-558: characterization of how Parkinson's disease mutants affect kinase activity. Biochem J 2007;405:307-317.
- Chan D, Citro A, Cordy JM, Shen GC, Wolozin B. Rac1 protein rescues neurite retraction caused by G2019S leucine-rich repeat kinase 2 (LRRK2). J Biol Chem 2011;286:16140-16149.
- Heo HY, Kim KS, Seol W. Coordinate regulation of neurite outgrowth by LRRK2 and its interactor, Rab5. Exp Neurobiol 2010;19:97-105.
- Chang F, Lemmon CA, Park D, Romer LH. FAK potentiates Rac1 activation and localization to matrix adhesion sites: a role for betaPIX. Mol Biol Cell 2007;18:253-264.
- Leu TH, Maa MC. Tyr-863 phosphorylation enhances focal adhesion kinase autophosphorylation at Tyr-397. Oncogene 2002;21:6992-7000.
- Calalb MB, Polte TR, Hanks SK. Tyrosine phosphorylation of focal adhesion kinase at sites in the catalytic domain regulates kinase activity: a role for Src family kinases. Mol Cell Biol 1995;15:954-963.
- Deramaudt TB, Dujardin D, Hamadi A, Noulet F, Kolli K, De Mey J, Takeda K, Rondé P. FAK phosphorylation at Tyr-925 regulates cross-talk between focal adhesion turnover and cell protrusion. Mol Biol Cell 2011;22:964-975.
- Calalb MB, Zhang X, Polte TR, Hanks SK. Focal adhesion kinase tyrosine-861 is a major site of phosphorylation by Src. Biochem Biophys Res Commun 1996;228:662-668.
- Abu-Ghazaleh R, Kabir J, Jia H, Lobo M, Zachary I. Src mediates stimulation by vascular endothelial growth factor of the phosphorylation of focal adhesion kinase at tyrosine 861, and migration and anti-apoptosis in endothelial cells. Biochem J 2001;360:255-264.
- Eliceiri BP, Puente XS, Hood JD, Stupack DG, Schlaepfer DD, Huang XZ, Sheppard D, Cheresh DA. Src-mediated coupling of focal adhesion kinase to integrin alpha(v)beta5 in vascular endothelial growth factor signaling. J Cell Biol 2002;157:149-160.
- Xie Z, Sanada K, Samuels BA, Shih H, Tsai LH. Serine 732 phosphorylation of FAK by Cdk5 is important for microtubule organization, nuclear movement, and neuronal migration. Cell 2003;114:469-482.



