Articles
Article Tools
Stats or Metrics
Article
Original Article
Exp Neurobiol 2019; 28(5): 628-641
Published online October 31, 2019
https://doi.org/10.5607/en.2019.28.5.628
© The Korean Society for Brain and Neural Sciences
LGR5 and Downstream Intracellular Signaling Proteins Play Critical Roles in the Cell Proliferation of Neuroblastoma, Meningioma and Pituitary Adenoma
Mina Hwang1†, Myung-Hoon Han2†, Hyun-Hee Park1*, Hojin Choi1, Kyu-Yong Lee1, Young Joo Lee1, Jae Min Kim2, Jin Hwan Cheong2, Je Il Ryu2, Kyueng-Whan Min3, Young-Ha Oh3, Yong Ko4 and Seong-Ho Koh1,5*
Departments of 1Neurology, 2Neurosurgery, and 3Pathology, Hanyang University Guri Hospital, Guri 11923, 4Department of Neurosurgery, Hanyang University Medical Center, Seoul 04763, 5Department of Translational Medicine, Hanyang University Graduate School of Biomedical Science & Engineering, Seoul 04763, Korea
Correspondence to: *To whom correspondence should be addressed.
Seong-Ho Koh, TEL: 82-31-560-2260, FAX: 82-31-560-2267
e-mail: ksh213@hanyang.ac.kr
Hyun-Hee Park, TEL: 82-31-560-2260, FAX: 82-31-560-2267
e-mail: newdiaz@hanmail.net
†These authors contributed equally to the work.
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License
(http://creativecommons.org/licenses/by-nc/4.0) which permits unrestricted non-commercial use, distribution, and
reproduction in any medium, provided the original work is properly cited.
Abstract
Leucine-rich repeat-containing G-protein coupled receptor 5 (LGR5) has been reported to play critical roles in the proliferation of various cancer cells. However, the roles of LGR5 in brain tumors and the specific intracellular signaling proteins directly associated with it remain unknown. Expression of LGR5 was first measured in normal brain tissue, meningioma, and pituitary adenoma of humans. To identify the downstream signaling pathways of LGR5, siRNA-mediated knockdown of
Graphical Abstract
Keywords: LGR5, Neuroblastoma, Meningioma, Pituitary adenoma, hnRNP
INTRODUCTION
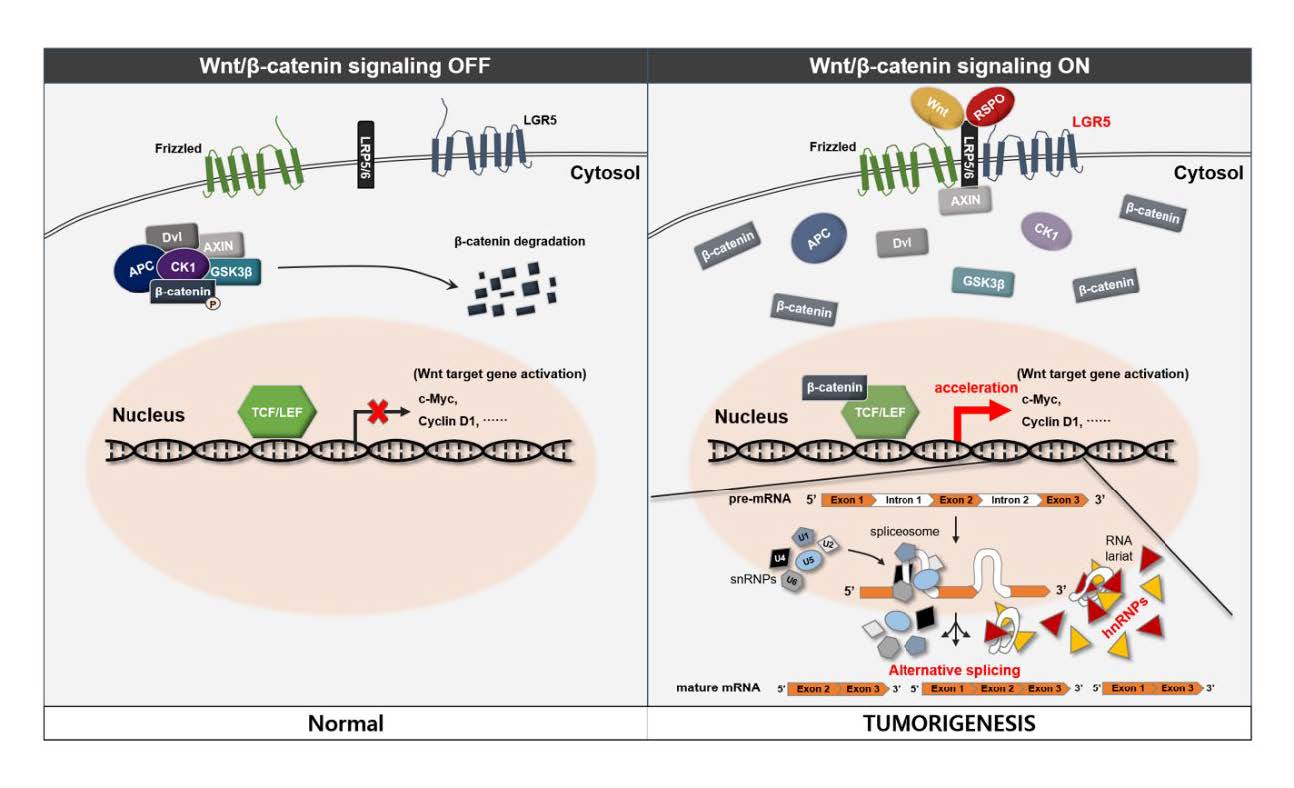
Cell surface receptor leucine-rich repeat-containing G-protein coupled receptor 5 (LGR5) is known as a stem cell marker in various organs and tissues including the gut, stomach, hair follicles, mammary gland, kidney, ovary, and neural tissue [1, 2]. LGR5 is a receptor for the R-spondin family of ligands and R-spondin-mediated activation of LGR5 promotes Wnt/β-catenin signaling. This process contributes to stem cell proliferation and self-renewal [3].
Wnt/β-catenin signaling is also known to be associated with meningioma and pituitary adenoma [4]. Although a recent study reported that LGR5 was significantly overexpressed in pituitary adenomas compared to normal pituitaries, to the best of our knowledge, there are no further studies regarding the association between LGR5 and meningioma and pituitary adenoma [5]. Therefore, we wanted to identify the possible association between LGR5, as well as its downstream signaling proteins, and meningioma or pituitary adenoma tissue in comparison to normal brain tissue. Knockdown of
Neuroblastoma is the most common extracranial solid tumor in childhood cancer. It arises from neural crest progenitor cells and can develop anywhere along the sympathetic nervous system [6]. LGR5 is highly expressed in high-grade neuroblastomas, and LGR5-activated Wnt/β-catenin signaling has been reported to play a critical role in neuroblastoma cell proliferation [7]. Indeed, short-interfering RNA (siRNA)-mediated knockdown of
Thus, the present study aimed to investigate whether the expression of LGR5 was different in meningioma and pituitary tumors compared with normal brain tissue in humans and to reveal proteins associated with LGR5 through siRNA-mediated knockdown of
MATERIALS AND METHODS
Preparation of human brain tumor tissues
Human brain whole tissue lysates and brain tissue slides from normal adults were obtained from Novus Biologicals (Littleton, CO, USA). The brain tumor tissue samples were obtained from patients who underwent surgical resection of meningioma and pituitary adenoma at the department of neurosurgery of Hanyang University Medical Center (Seoul and Guri), Korea, from November 2016 in Guri and March 2017 in Seoul. Resected fresh tumor tissues were collected during surgery, these samples were then immediately submitted to the laboratory for storage at −80°C in a facility which is located very near the operation room.
The study protocol was reviewed and approved by the Institutional Review Board in both Seoul (IRB No. 2017-02-016) and Guri Hospitals (IRB No. 2016-10-002) and adhered to the tenets of the Declaration of Helsinki. All patients provided informed written consent prior to participation in this study.
MRI acquisition and histological sample preparation for light microscopy
All brain MRI images were acquired using Philips 3.0 Tesla MRI scanners (Ingenia CX and Achieva, Philips Medical Systems, Best, The Netherlands) in both hospitals.
Fresh tumor tissues were fixed in 10% formalin for 24 h, then grossed and placed for processing in an automated tissue processor (Thermo Fisher Scientific, Sydney, Australia). The tissue sections were sliced at 5 μm thickness using a microtome. Hematoxylin and eosin (H&E) stains were performed using a Tissue-Tek Prisma® E2D automated slide stainer (Sakura Finetek Japan Co., Ltd., Tokyo, Japan), following the respective standard protocols. All histopathological diagnoses established according to the World Health Organization (WHO) classification were additionally reviewed by two pathologists (K.W.M, Y.H.O) [10, 11].
Cell culture of SH-SY5Y cells
SH-SY5Y cells were obtained from the Korean Cell Line Bank (KCLB). Cells were plated on culture dishes and cultured in MEM medium [1× Minimum Essential Medium (MEM, Gibco, Frederick, MD, USA), 25 mM HEPES, 25 mM sodium bicarbonate (Sigma-Aldrich, St. Louis, MO, USA), 90%; heat-inactivated fetal bovine serum (FBS, Gibco), 10%]. The media was changed every 3~4 days, and cultures were maintained at 37°C under a humidified 5% CO2 atmosphere.
LGR5 knockdown
SH-SY5Y cells were seeded at a density of 2.5×106 cells/cm2 in two six-well culture plates and cultured in antibiotic-free normal growth medium supplemented with FBS. Cells were incubated up to 60~80% confluency for 18~24 h and
Western blot analysis
Expression levels of LGR5, active β-catenin, cyclin D1, c-myc, hnRNPA2B1, and hnRNPH3 in human normal and collected brain tumor tissues and knockdown of LGR5 in SH-SY5Y cells were confirmed by western blot analysis [12, 13]. Briefly, sample tissues and cells were washed twice with cold D-PBS and incubated for 30 m on ice after the addition of lysis buffer (RIPA II cell lysis buffer 1× with Triton, without EDTA; 1 mM phenylmethyl-sulfonyl fluoride (PMSF); 1 mM sodium fluoride (NaF); 1 mM sodium orthovanadate (Na3VO4); and 0.5% protease inhibitor cocktail 1×). Next, the tissues and cells were sonicated (Sonoplus, Bandelin Electronics, Berlin, Germany) and incubated for 30 m on ice. The resulting lysates were then centrifuged at 16,200×g for 15 m and the protein concentrations of lysates were determined using a Bio-Rad (Hercules, CA, USA) protein assay kit. Normal adult human brain whole lysates (Novus Biologicals) and lysate samples containing equal amounts of protein (tissues, 30 μg; cells, 25 μg) were resolved by 4~12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE, Invitrogen). After SDS-PAGE, proteins were transferred to polyvinylidene fluoride membranes (Millipore, Bedford, MA, USA). Membranes were blocked with 2% skimmed milk and incubated with a specific primary antibody. The antibodies used in these experiments were as follows: LGR5 (1:200, Santa Cruz), active β-catenin (1:1000, Cell Signaling, Beverly, MA, USA), cyclin D1 (1:2000, Cell Signaling), c-myc (1:1000, Cell Signaling), GAPDH (1:4000, Cell Signaling), hnRNPA2B1 (0.75 μg/ml, Abcam, Burlingame, CA, USA), and hnRNPH3 (1:75, Santa Cruz). After washing membranes three times with Tris-buffered saline containing 0.1% Tween-20 (TBST), membranes were hybridized with Horseradish Peroxidase (HRP)-conjugated anti-mouse antibody (1:2000, Jackson ImmunoResearch Laboratories, Inc., West Grove, PA, USA) or HRP-conjugated anti-rabbit antibody (1:2000, Jackson ImmunoResearch Laboratories) followed by using a West-Q Chemiluminescent Substrate Kit (GenDEPOT, Katy, TX, USA). Blots were quantified with an image analyzer (ImageQuant LAS 4000; GE Healthcare, Little Chalfont, UK).
Immunohistochemistry
For immunohistochemistry, human brain normal and tumor tissues (meningioma and pituitary adenoma) were fixed with paraffin. First, fixed tissues were subjected to deparaffinization and hydration processes. The tissues were washed with xylene 4 times, 100% ethanol 2 times, 95% ethanol, 80% ethanol, 70% ethanol, 50% ethanol, and auto-distilled water for 3 m each step. Antigen retrieval was induced from deparaffinized and hydrated tissues using pre-heated 0.01 M sodium citrate (pH 6). The tissues were permeabilized with 0.5% Triton X-100 (Sigma-Aldrich) in 0.01 M PBS for 5 m. Next, H2O2 was used to block endogenous peroxidase activity of the tissues for 10 m and 10% FBS (Gibco) was used as the blocking solution for 1 h. The tissues were then incubated overnight with specific primary antibodies in 0.5% FBS at 4°C. The antibodies used in these experiments were hnRNPH3 (1:300, Santa Cruz) and LGR5 (1:60, Abcam). The following day, the tissues were washed and incubated with secondary antibodies in 0.5% FBS for 3 h. Tetramethylrhodamine goat anti-mouse IgG (Invitrogen) and Alexa FluorTM 488 goat anti-rabbit IgG (Invitrogen) were used as secondary antibodies. Finally, the tissues were mounted with DAPI mounting solution (Vector Laboratories Inc., Burlingame, CA, USA). Throughout the procedure, the tissues were washed three times with 0.01 M PBS between each step. Immunohistochemically positive cells were identified under a confocal fluorescence microscope (Nanoscope systems Inc., Daejeon, Korea).
Proteomics
SH-SY5Y cells treated with LGR5 or control siRNA were collected and washed several times with D-PBS to remove salts and lipids. The cells were then lysed in lysis buffer consisting of 7 M urea and 2 M thiourea containing 4%(w/v) 3-[(3-cholamidopropyl) dimethylammonio]-1-propanesulfonate (CHAPS), 1% (w/v) di-thiothreitol (DTT), 2% (v/v) pharmalyte, and 1 mM benzamidine. The cells were then sonicated as described above and extracted by vortexing at room temperature for 1 h. Next, the lysates were subjected to centrifugation at 15,000×g for 1 h at 4°C, after which the insoluble material was discarded and the soluble fraction was saved for two-dimensional gel electrophoresis (2-DE). Protein concentrations were calculated by using the Bradford assay.
Protein samples prepared as described above were added to 0.5% Immobilized pH gradient (IPG) buffer and re-hydrated with IPG dry strips (3~10 NL IPG, 13 cm, GE Healthcare, Buckinghamshire, UK) for 16~20 h. Next, isoelectric focusing (IEF) was performed on the re-hydrated samples using an Ettan IPGphor 3 (GE Healthcare) according to the manufacturer’s instructions. During IEF, the voltage was increased to 8,000 V over 7 h, followed by maintenance at a constant 8,000 V for sample entry, and focusing was completed after 55 kV. Prior to the 2nd dimension separation, the isoelectric-focused strips were incubated in equilibration buffer (75 mM Tris-Cl, pH 8.8, containing 6 M urea, 2% SDS, 0.002% 1% Bromophenol blue stock solution, and 29.3% glycerol) containing 1% DTT (Sigma-Aldrich) for 15 m and then the same procedure was repeated with equilibration buffer containing 2.5% iodoacetamide (Sigma-Aldrich) for 15 m. The equilibrated strips were then fixed onto large SDS-PAGE gels (13×18 cm, 12.5%) using an agarose sealing solution. 2D-PAGE was performed using a SE600 2D system (GE Healthcare) according to the manufacturer’s instructions. Briefly, 2D gels were run at 4°C 10 mA for 1.5 h, 30 mA for 3 h, and 40 mA for 2 h.
Completed 2D-PAGE gels were stained using Coomassie brilliant blue R-350 (CBB; GE Healthcare) and scanned as computer images. The collected images were quantitatively analyzed by ImageMasterTM 2D Platinum 7.0 software (GE Healthcare) according to the manufacturer’s instructions. The intensity of each protein spot was normalized to the total intensity of all validated protein spots. The expression of the selected protein spots had at the least a two-fold significant difference compared to control samples.
Proteins were identified by peptide mass fingerprinting (PMF). Protein spots were excised, digested with trypsin (Promega, Madison, WI, USA), and mixed with α cyano-4-hydroxycinnamic acid in 50% acetonitrile/0.1% TFA for MALDI-TOF analysis (Microflex LRF 20, BrukerDaltonics, Billerica, MA, USA) as described by Fernandez et al [14]. Spectra were collected from 300 shots per spectrum over the 600~3000 m/z range and calibrated by a two-point internal calibration using trypsin auto-digestion peaks (m/z 842.5099, 2211.1046). The peak list was generated by Flex Analysis 3.0 software, with a peak selection threshold of 500 for the minimum resolution of monoisotopic mass and 5 for S/N. Protein identification by PMF was performed using the search program MASCOT, developed by Matrix Science (
Bioinformatics analysis
A network of associated proteins was constructed using the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database version 11.0 (
Statistical analysis
All data are expressed as the means±SD of three or more independent experiments. Statistical comparisons between different treatment groups were performed with the Student’s t-test and one-way analysis of variance (ANOVA) followed by Tukey’s honestly significant difference (HSD) test. p-values less than 0.05 were considered statistically significant.
RESULTS
Knockdown of LGR5 in SH-SY5Y cells
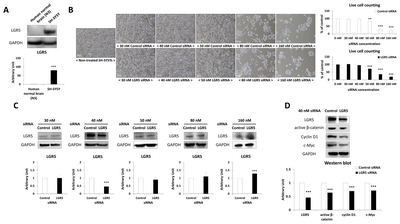
SH-SY5Y cells showed significantly higher LGR5 expression compared to human normal brain tissue (Fig. 1A). Different concentrations of
Identification of differentially regulated proteins after LGR5 knockdown in SH-SY5Y cells
Proteomics was performed using 2D-PAGE to evaluate differences in the expression of intracellular proteins in neuroblastoma cells with or without
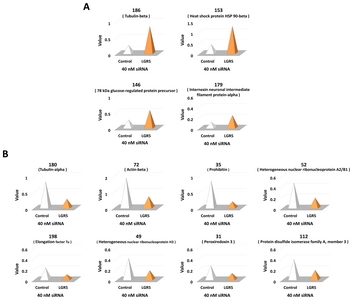
Multiple landmarks were selected for the classification of protein spots. Based on the landmarks, 12 protein spots were identified that exhibited a greater than two-fold increase or decrease in expression in SH-SY5Y cells treated with
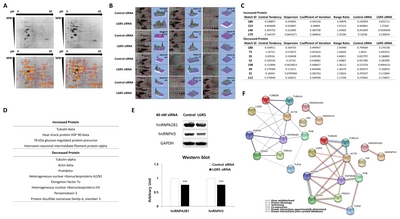
Identification of proteins altered by LGR5 knockdown using MALDI-TOF mass spectrometry
A total of 12 protein spots altered by
Among the 12 proteins identified, the expression of four proteins was increased in neuroblastoma cells with
Protein-protein interactions associated with LGR5 in neuroblastoma cells
Direct or indirect interactions with specific interaction types were identified among the 12 proteins that were associated with LGR5 in neuroblastoma cells (Fig. 2F). As expected, a strong protein interaction was observed between each protein family, which included cytoskeleton proteins, HSPs, and hnRNPs. Interestingly, the HSP family exhibited strong protein interactions with cytoskeleton proteins and PDIA3. In addition, peroxiredoxin 3 was found to interact strongly with PDIA3. However, due to the lack of previous information on proteins associated with LGR5, especially in neuroblastoma cells, no significant relationships between LGR5 and the 12 differentially expressed proteins were identified.
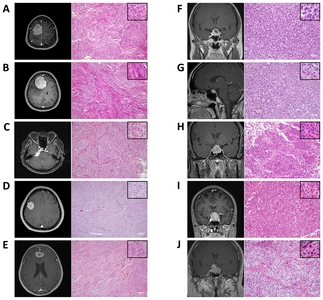
Clinical, radiographic, and histologic characteristics of study subjects
In the present study, we included three human tissues from normal brain, five benign meningiomas (WHO grade I), and five pituitary adenomas. Detailed information of patients and tumor samples is presented in Table 1.
Representative radiographic and histological findings of meningothelial, fibrous, and transitional meningioma, and pituitary adenoma in study subjects are shown in Fig. 4.
LGR5 expression in human brain tumor tissue samples
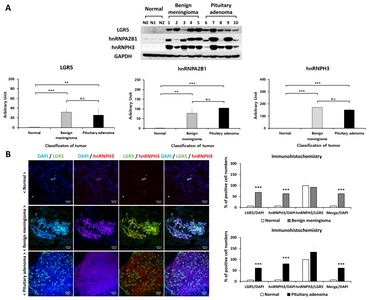
LGR5 expression was measured in normal brain, benign meningioma, and pituitary adenoma tissues. Fig. 5A shows differences of LGR5 protein expression, analyzed by western blotting, between normal, benign meningioma, and pituitary adenoma. It was found that LGR5 expression was significantly higher in meningioma and pituitary adenoma, compared to normal brain tissue.
Expression of LGR5 and hnRNP proteins in meningioma and pituitary adenoma tissues
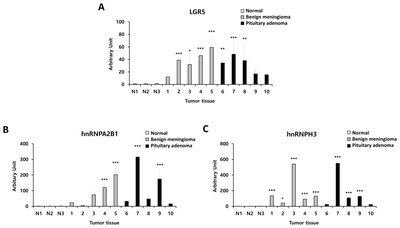
We also evaluated hnRNPA2B1 and hnRNPH3 expression levels in benign meningioma and pituitary adenoma tissues compared to normal brain tissue by using western blotting (Fig. 5A). We identified significantly higher expression of hnRNPA2B1 and hnRNPH3 in benign meningioma and pituitary adenoma compared to normal. Detailed information on protein expression levels of LGR5, hnRNPA2B1, and hnRNPH3 in each meningioma and pituitary adenoma tissue sample compared to normal is presented in Fig. 6.
The distribution of LGR5 and hnRNPH3 was identified in human normal brain and benign meningioma and pituitary adenoma with immunohistochemical analysis (Fig. 5B). We observed that LGR5 and hnRNPH3 were co-expressed in meningioma and pituitary adenoma, indicating that both LGR5 and hnRNPs are associated with human meningioma and pituitary adenoma.
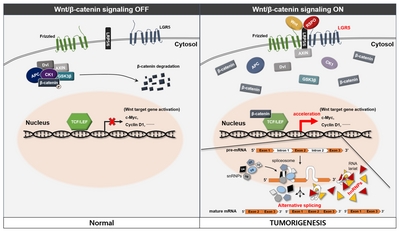
Proposed mechanisms for the role of LGR5 and hnRNPs in human meningioma and pituitary adenoma
In considering our results, we propose a possible novel association between LGR5 and the hnRNP family in human meningioma and pituitary adenoma (Fig. 7). The detailed mechanisms of Wnt signaling and splicing, which were not proved in the study, were referenced from previous studies [16–19]. R-spondin-mediated activation of LGR5 promotes Wnt/β-catenin signaling and the activation of Wnt target genes may also affect activation of the hnRNP family. This process may lead to alternative gene transcription, splicing, and post-transcriptional modification of pre-mRNA processing and result in tumorigenesis in human meningioma and pituitary adenoma.
DISCUSSION
A few previous studies reported that LGR5 was related to poor prognosis or proliferation of intra-axial brain malignancy such as glioma [20, 21]. However, to the best of our knowledge, there is a lack of studies regarding the association between LGR5 and extra-axial brain tumors such as meningioma and pituitary adenoma. In addition, LGR5 is closely associated with Wnt signaling, and previous studies have reported that both meningioma and pituitary adenoma are associated with Wnt signaling [8, 9]. Therefore, we wanted to investigate the association between LGR5 and human meningioma and pituitary adenoma and confirmed that LGR5 was increased in meningioma and pituitary tumors compared with normal brain tissue (Fig. 4 and 5A). Initially, we evaluated the proteins which are associated with
In the present study, we initially identified 12 proteins in the SH-SY5Y neuroblastoma cell line that were differentially expressed following knockdown of
Activation of the Wnt/β-catenin pathway can upregulate c-myc expression [23], and a recent study suggested that hnRNP-K is a potential downstream signaling protein involved in the Wnt/c-Myc pathway [24]. Therefore, we hypothesized that down-regulation of Wnt/β-catenin signaling by
We also found that
Lastly, we found that expression of α-internexin was increased by
There are limitations associated with the study. First, the initial utilization of neuroblastoma cells instead of using meningioma or pituitary adenoma cell lines may limit our findings. However, several clinical reports showed possible associations between neuroblastoma, and meningioma and pituitary adenoma. A previous study reported meningioma development after radiotherapy for an olfactory neuroblastoma at the same site [34]. The authors described that growth factor/cytokine release (such as insulin-like growth factor 1) by olfactory neuroblastoma has been linked to the oncogenesis of meningiomas [35–37]. In addition, previous studies have reported cases of neuroblastoma arising in the sellar region, and suggested that it mimicked a non-functioning pituitary adenoma [38–41]. Second, LGR5 knockdown with siRNA in SH-SY5Y neuroblastoma cells showed only approximately 50% efficiency, and this relatively low knockdown efficiency of LGR5 may affect the results. However, several studies have also reported a similar siRNA knockdown efficiency [42–45]. Third, there were significant differences in the LGR5 expression between each tumor sample. However, the tumor samples showing relatively low LGR5 expressions showed higher LGR5 expressions compared to normal brain samples.
In conclusion, we identified a total of 12 proteins whose expression was altered by
ACKNOWLEDGEMENTS
This research was supported by the Basic Science Research Program of the National Research Foundation of Korea, which is funded by the Ministry of Science, ICT, and Future Planning (2018R1D1A1B07047722, 2018R1A2A2A15023219, and 2018R1D1A1A09082825); funded by the Ministry of Health & Welfare, Republic of Korea (grant number: HI18C1254), and by the Medical Research Center (2017R1A5A2015395).
Figures
Tables
Table 1 Patient description and tumor sample characteristics
| Sample | Sex | Age | Tumor type | Location | Sample Concentration (μg/μl) | Sample volume (μl) | Ki-67 index | p53 | Underlying disease |
|---|---|---|---|---|---|---|---|---|---|
| N1 | M | 66 | Normal brain (human adult) | Whole brain | 4.47 | 150 | |||
| N2 | M | 57 | Normal brain (human adult) | Whole brain | 4.32 | 150 | |||
| N3 | M | 82 | Normal brain (human adult) | Whole brain | 4.89 | 150 | |||
| Group 1 | Meningioma, WHO grade I | ||||||||
| 1 | M | 61 | Meningioma, meningothelial | Parasagittal, right | 28.62 | 110 | <1% | (−) | Alcohol, smoking |
| 2 | F | 64 | Meningioma, fibrous | Falx, right | 29.13 | 200 | 1~2% | (−) | (−) |
| 3 | F | 64 | Meningioma, meningothelial | CPA, right | 23.01 | 100 | 1% | (−) | (−) |
| 4 | F | 56 | Meningioma, transitional | Convexity, right | 41.67 | 600 | <1% | 15% | HTN, DM |
| 5 | F | 74 | Meningioma, transitional | Falx, both | 30.99 | 400 | <1% | 30% | (−) |
| Group 2 | Pituitary adenoma | ||||||||
| 6 | F | 51 | Pituitary adenoma | Pituitary gland | 19.47 | 100 | <1% | 1% | (−) |
| 7 | F | 42 | Pituitary adenoma, ACTH producing | Pituitary gland | 17.94 | 90 | <1% | 1% | Cushing disease |
| 8 | M | 51 | Pituitary adenoma | Pituitary gland | 20.19 | 110 | <1% | (−) | DM |
| 9 | M | 38 | Pituitary adenoma | Pituitary gland | 20.31 | 50 | 1% | 1% | (−) |
| 10 | F | 53 | Pituitary adenoma | Pituitary gland | 18.00 | 90 | <1% | 5% | (−) |
WHO, world health organization; CPA, cerebellopontine angle; HTN, hypertension; DM, diabetes mellitus; ACTH, adrenocorticotropic hormone.
References
- Lin Y, Fang ZP, Liu HJ, Wang LJ, Cheng Z, Tang N, Li T, Liu T, Han HX, Cao G, Liang L, Ding YQ, Zhou WJ (2017) HGF/Rspondin1 rescues liver dysfunction through the induction of Lgr5+ liver stem cells. Nat Commun 8:1175.
- Satoh J, Obayashi S, Tabunoki H, Wakana T, Kim SU (2010) Stable expression of neurogenin 1 induces LGR5, a novel stem cell marker, in an immortalized human neural stem cell line HB1.F3. Cell Mol Neurobiol 30:415-426.
- Scannell CA, Pedersen EA, Mosher JT, Krook MA, Nicholls LA, Wilky BA, Loeb DM, Lawlor ER (2013) LGR5 is expressed by Ewing sarcoma and potentiates Wnt/β-catenin signaling. Front Oncol 3:81.
- Howng SL, Wu CH, Cheng TS, Sy WD, Lin PC, Wang C, Hong YR (2002) Differential expression of Wnt genes, beta-catenin and E-cadherin in human brain tumors. Cancer Lett 183:95-101.
- Li J, Zhang Y, Cheng N, Zhao Q, Liu H, Song S (2016) The overexpression of leucine-rich repeat-containing g-proteincoupled receptor 5 in pituitary adenomas. Transl Surg 1:37- 40.
- Maris JM, Hogarty MD, Bagatell R, Cohn SL (2007) Neuroblastoma. Lancet 369:2106-2120.
- Vieira GC, Chockalingam S, Melegh Z, Greenhough A, Malik S, Szemes M, Park JH, Kaidi A, Zhou L, Catchpoole D, Morgan R, Bates DO, Gabb PD, Malik K (2015) LGR5 regulates pro-survival MEK/ERK and proliferative Wnt/β-catenin signalling in neuroblastoma. Oncotarget 6:40053-40067.
- Gaston-Massuet C, Andoniadou CL, Signore M, Jayakody SA, Charolidi N, Kyeyune R, Vernay B, Jacques TS, Taketo MM, Le Tissier P, Dattani MT, Martinez-Barbera JP (2011) Increased Wingless (Wnt) signaling in pituitary progenitor/ stem cells gives rise to pituitary tumors in mice and humans. Proc Natl Acad Sci U S A 108:11482-11487.
- Pećina-Šlaus N, Kafka A, Lechpammer M (2016) Molecular genetics of intracranial meningiomas with emphasis on canonical Wnt signalling. Cancers (Basel) 8:E67.
- IARC International Agency for Research on Cancer (2017) WHO classification of tumours of endocrine organs. 4th ed. World Health Organization, Lyon.
- Louis DN, Ohgaki H, Wiestler OD, Cavenee WK (2016) WHO classification of tumours of the central nervous system. International Agency for Research On Cancer, Lyon.
- Hong EH, Hwang M, Shin YU, Park HH, Koh SH, Cho H (2018) Leucine-rich G protein-coupled receptor-5 is significantly increased in the aqueous humor of human eye with proliferative diabetic retinopathy. Exp Neurobiol 27:238-244.
- Lee J, Park HH, Koh SH, Choi H (2017) Neural stem cell death mechanisms induced by amyloid beta. Dement Neurocogn Disord 16:121-127.
- Fernandez J, Gharahdaghi F, Mische SM (1998) Routine identification of proteins from sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels or polyvinyl difluoride membranes using matrix assisted laser desorption/ ionization-time of flight-mass spectrometry (MALDI-TOFMS). Electrophoresis 19:1036-1045.
- Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, Kuhn M, Bork P, Jensen LJ, von Mering C. (2015) STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 43:D447-D452.
- Huelga SC, Vu AQ, Arnold JD, Liang TY, Liu PP, Yan BY, Donohue JP, Shiue L, Hoon S, Brenner S, Ares M Jr, Yeo GW (2012) Integrative genome-wide analysis reveals cooperative regulation of alternative splicing by hnRNP proteins. Cell Rep 1:167-178.
- Dvinge H, Kim E, Abdel-Wahab O, Bradley RK (2016) RNA splicing factors as oncoproteins and tumour suppressors. Nat Rev Cancer 16:413-430.
- Baron R, Kneissel M (2013) WNT signaling in bone homeostasis and disease: from human mutations to treatments. Nat Med 19:179-192.
- Lee Y, Lee JK, Ahn SH, Lee J, Nam DH (2016) WNT signaling in glioblastoma and therapeutic opportunities. Lab Invest 96:137-150.
- Wang D, Zhou J, Fan C, Jiao F, Liu B, Sun P, Miao J, Zhang Q (2014) Knockdown of LGR5 suppresses the proliferation of glioma cells in vitro and in vivo. Oncol Rep 31:41-49.
- Nakata S, Campos B, Bageritz J, Bermejo JL, Becker N, Engel F, Acker T, Momma S, Herold-Mende C, Lichter P, Radlwimmer B, Goidts V (2013) LGR5 is a marker of poor prognosis in glioblastoma and is required for survival of brain cancer stem-like cells. Brain Pathol 23:60-72.
- Sakka L, Delétage N, Chalus M, Aissouni Y, Sylvain-Vidal V, Gobron S, Coll G (2017) Assessment of citalopram and escitalopram on neuroblastoma cell lines. Cell toxicity and gene modulation. Oncotarget 8:42789-42807.
- Zhang S, Li Y, Wu Y, Shi K, Bing L, Hao J (2012) Wnt/β-catenin signaling pathway upregulates c-Myc expression to promote cell proliferation of P19 teratocarcinoma cells. Anat Rec (Hoboken) 295:2104-2113.
- Kawasaki Y, Komiya M, Matsumura K, Negishi L, Suda S, Okuno M, Yokota N, Osada T, Nagashima T, Hiyoshi M, Okada-Hatakeyama M, Kitayama J, Shirahige K, Akiyama T (2016) MYU, a target lncRNA for Wnt/c-Myc signaling, mediates induction of CDK6 to promote cell cycle progression. Cell Rep 16:2554-2564.
- Deng J, Chen S, Wang F, Zhao H, Xie Z, Xu Z, Zhang Q, Liang P, Zhai X, Cheng Y (2016) Effects of hnRNP A2/B1 knockdown on inhibition of glioblastoma cell invasion, growth and survival. Mol Neurobiol 53:1132-1144.
- Liang Y, Shi SL, Li QF, Chen LY, Jing GJ, Tan GW, Wang SY, Wu FY (2011) The localization of hnRNP A2/B1 in nuclear matrix and the aberrant expression during the RA-induced differentiation of human neuroblastoma SK-N-SH cells. J Cell Biochem 112:1722-1729.
- Wang E, Aslanzadeh V, Papa F, Zhu H, de la Grange P, Cambi F (2012) Global profiling of alternative splicing events and gene expression regulated by hnRNPH/F. PLoS One 7:e51266.
- Shilo A, Ben Hur V, Denichenko P, Stein I, Pikarsky E, Rauch J, Kolch W, Zender L, Karni R (2014) Splicing factor hnRNP A2 activates the Ras-MAPK-ERK pathway by controlling ARaf splicing in hepatocellular carcinoma development. RNA 20:505-515.
- Zhang S, Wei JS, Li SQ, Badgett TC, Song YK, Agarwal S, Coarfa C, Tolman C, Hurd L, Liao H, He J, Wen X, Liu Z, Thiele CJ, Westermann F, Asgharzadeh S, Seeger RC, Maris JM, Guidry Auvil JM, Smith MA, Kolaczyk ED, Shohet J, Khan J (2016) MYCN controls an alternative RNA splicing program in high-risk metastatic neuroblastoma. Cancer Lett 371:214-224 .
- Wohlgemuth I, Pohl C, Mittelstaet J, Konevega AL, Rodnina MV (2011) Evolutionary optimization of speed and accuracy of decoding on the ribosome. Philos Trans R Soc Lond B Biol Sci 366:2979-2986.
- Defeu Soufo HJ, Reimold C, Breddermann H, Mannherz HG, Graumann PL (2015) Translation elongation factor EF-Tu modulates filament formation of actin-like MreB protein in vitro. J Mol Biol 427:1715-1727.
- Nishinaka Y, Masutani H, Nakamura H, Yodoi J (2001) Regulatory roles of thioredoxin in oxidative stress-induced cellular responses. Redox Rep 6:289-295.
- Zhao J, Liem RKH (2016) α-internexin and peripherin: expression, assembly, functions, and roles in disease. In: Methods in enzymology (Omary MB, Liem RKH, eds), pp 477-507. Academic Press, Cambridge, MA.
- Johnson MD, Piech K, Emandian S (2013) Radiation-associated meningioma in the elderly: development of meningioma with olfactory neuroblastoma recurrence 10 years after irradiation. Ann Clin Lab Sci 43:460-463.
- Sandberg-Nordqvist AC, Ståhlbom PA, Reinecke M, Collins VP, von Holst H, Sara V (1993) Characterization of insulinlike growth factor 1 in human primary brain tumors. Cancer Res 53:2475-2478.
- Yee D, Favoni RE, Lebovic GS, Lombana F, Powell DR, Reynolds CP, Rosen N (1990) Insulin-like growth factor I expression by tumors of neuroectodermal origin with the t(11;22) chromosomal translocation. A potential autocrine growth factor. J Clin Invest 86:1806-1814.
- Johnson M, Toms S (2005) Mitogenic signal transduction pathways in meningiomas: novel targets for meningioma chemotherapy? J Neuropathol Exp Neurol 64:1029-1036.
- Whee SM, Lee JI, Kim JH (2002) Intrasellar schwannoma mimicking pituitary adenoma: a case report. J Korean Med Sci 17:147-150.
- Dupuy M, Bonneville F, Grunenwald S, Breibach F, Delisle MB, Chaynes P, Sol JC, Caron P (2012) Primary sellar neuroblastoma. A new case and review of literature. Ann Endocrinol (Paris) 73:216-221.
- Schmalisch K, Psaras T, Beschorner R, Honegger J (2009) Sellar neuroblastoma mimicking a pituitary tumour: case report and review of the literature. Clin Neurol Neurosurg 111:774-778.
- Oyama K, Yamada S, Usui M, Kovacs K (2005) Sellar neuroblastoma mimicking pituitary adenoma. Pituitary 8:109-114.
- Prechtel AT, Turza NM, Theodoridis AA, Steinkasserer A (2007) CD83 knockdown in monocyte-derived dendritic cells by small interfering RNA leads to a diminished T cell stimulation. J Immunol 178:5454-5464.
- Kobayashi M, Chandrasekhar A, Cheng C, Martinez JA, Ng H, de la Hoz C, Zochodne DW (2017) Diabetic polyneuropathy, sensory neurons, nuclear structure and spliceosome alterations: a role for CWC22. Dis Model Mech 10:215-224.
- Shi YA, Zhao Q, Zhang LH, Du W, Wang XY, He X, Wu S, Li YL (2014) Knockdown of hTERT by siRNA inhibits cervical cancer cell growth in vitro and in vivo. Int J Oncol 45:1216-1224.
- Bruneau S, Néel M, Roumenina LT, Frimat M, Laurent L, Frémeaux-Bacchi V, Fakhouri F (2015) Loss of DGKε induces endothelial cell activation and death independently of complement activation. Blood 125:1038-1046.





