Articles
Article Tools
Stats or Metrics
Article
Original Article
Exp Neurobiol 2016; 25(1): 14-23
Published online February 29, 2016
https://doi.org/10.5607/en.2016.25.1.14
© The Korean Society for Brain and Neural Sciences
PINK1 Deficiency Decreases Expression Levels of mir-326, mir-330, and mir-3099 during Brain Development and Neural Stem Cell Differentiation
Insup Choi1,2, Joo Hong Woo2, Ilo Jou1,2,3 and Eun-hye Joe1,2,3,4,5*
1Neuroscience Graduate Program Department of Biomedical Sciences, 2Chronic Inflammatory Disease Research Center, 3Department of Pharmacology, 4Department of Brain Science, 5Brain Disease Research Center, Ajou University School of Medicine, Suwon 16499, Korea
Correspondence to: *To whom correspondence should be addressed.
TEL: 82-31-219-5062, FAX: 82-31-219-5069
e-mail: ehjoe@ajou.ac.kr
PTEN-induced putative kinase 1 (PINK1) is a Parkinson's disease (PD) gene. We examined miRNAs regulated by PINK1 during brain development and neural stem cell (NSC) differentiation, and found that lvels of miRNAs related to tumors and inflammation were different between 1-day-old-wild type (WT) and PINK1-knockout (KO) mouse brains. Notably, levels of miR-326, miR-330 and miR-3099, which are related to astroglioma, increased during brain development and NSC differentiation, and were significantly reduced in the absence of PINK1. Interestingly, in the presence of ciliary neurotrophic factor (CNTF), which pushes differentiation of NSCs into astrocytes, miR-326, miR-330, and miR-3099 levels in KO NSCs were also lower than those in WT NSCs. Furthermore, mimics of all three miRNAs increased expression of the astrocytic marker glial fibrillary acidic protein (GFAP) during differentiation of KO NSCs, but inhibitors of these miRNAs decreased GFAP expression in WT NSCs. Moreover, these miRNAs increased the translational efficacy of GFAP through the 3'-UTR of GFAP mRNA. Taken together, these results suggest that PINK1 deficiency reduce expression levels of miR-326, miR-330 and miR-3099, which may regulate GFAP expression during NSC differentiation and brain development.
Keywords: PINK1, Parkinson’s disease, miR-326, miR-330, miR-3099
Parkinson's disease (PD), the second-most common neurodegenerative disease, is accompanied by dopaminergic neuronal death in the substantia nigra pars compacta. Although most PD cases are sporadic, considerable effort has been devoted to studying the function of PD-associated genes to gain insights into the onset and progression of PD.
PTEN-induced putative kinase 1 (PINK1) is a PD-related gene that, when mutated, causes an autosomal recessive, early-onset PD [1]. PINK1 has pro-survival, anti-apoptotic and cytoprotective functions [2]. PINK1 regulates ATP generation, oxygen consumption [3,4,5], and reactive oxygen species (ROS) production [6] through regulation of mitochondrial function [7,8,9,10]. In addition, PINK1 acts through the AKT-mTOR pathway to regulate survival, proliferation, metabolism, and inflammation, among other cellular functions [11,12,13,14,15,16].
MicroRNAs (miRNAs) are small, noncoding RNA molecules that influence diverse biological functions, including brain development and diseases, through regulation of mRNA and protein expression. Expression of miRNAs is highly dynamic, and miRNAs are crucial for proper neuronal and glial differentiation during brain development [17,18,19,20,21]. Accordingly, deregulation of miRNAs has been reported in several types of neurodegenerative diseases [22,23,24,25,26,27,28,29,30,31,32,33,34,35,36]. In the brain of Alzheimer's disease patients and aged individuals, miRNAs that regulate expression of β-site amyloid precursor protein (APP)-cleaving enzyme (BACE), including miR-107, miR-298 and miR-328, are decreased [25,26]. In the case of Huntington's disease, expression of miR-9/9*, which targets REST (repressor element 1 silencing transcription factor), is also decreased [27]. In multiple sclerosis, miR-155 and miR-326 are increased, but miR-15a, miR-15b, miR-181c and miR-328 are down-regulated [28]. miRNAs are also linked to PD. In the adult midbrain, specific depletion of Dicer, an endoribonuclease necessary for maturation of miRNAs, causes degeneration of dopaminergic neurons, suggesting that miRNAs maintain dopaminergic neurons in adulthood. Expression of miR-133b, which regulates maturation of dopaminergic neurons, is downregulated in PD patients [29]. miR-9 and miR-326 regulate human dopamine D2 receptor expression [30]. The G2019S mutant of LRRK2 (leucine-rich repeat kinase 2) inhibits the functions of let-7 and miR-184*, which cause neuronal toxicity by forcing cell-cycle progression in post-mitotic neurons [31]. Recently, miR-330 has been identified as a novel gene that is related to PD [32]. In addition, miR-7 and miR-153 have been shown to suppress expression of α-synuclein, a major component of Lewy bodies in PD patients [33,34,35], and miR-155 has been shown to mediate the anti-inflammatory effects of DJ-1 (PARK1), a PD-related gene [36]. In Drosophila, pathogenic G2019S and I1915T mutants of LRRK2 antagonize let 7 and miR-184*, and induce death of dopaminergic neurons [31].
In this study, we show that expression of miRNAs, particularly miR-326, miR-330 and miR-3099 increases during brain development and neural stem cell (NSC) differentiation. Interestingly, PINK1 deficiency decreases expression of these miRNAs during these processes. Collectively, these findings suggest that defects in the expression of these miRNAs could be linked to the development of PD caused by a PINK1 deficiency.
MATERIALS AND METHODS
PINK1-deficient mice were a generous gift from Drs. Zhuang (Chicago University) and Kang (Columbia University). PINK1-deficient mice were generated by replacing a 5.6-kb genomic region of the PINK1 locus, including exons 4~7 and the coding portion of exon 8, with a PGK-neo-polyA selection cassette flanked by FRT sequences, as previously described [37]. Heterozygous mice were bred to generate PINK1-null mice and their WT littermate controls for experiments. Mice were genotyped by multiplex PCR of genomic DNA extracted from tail snips. The first primer pair amplified part of intron 6 of PINK1 (present in all mice); the second primer pair amplified part of neo (absent in WT mice); and the third primer pair amplified intron 6 of PINK1 (absent in homozygous mutants). The absence of PINK1 expression was verified by reverse transcription-PCR (RT-PCR) and in situ hybridization (data not shown). All animal procedures were approved by the Ajou University School of Medicine Ethics Review Committee for Animal Research (Amc-119).
Embryonic neurospheres were cultured from E13.5 mouse brains as previously described [38]. Briefly, forebrains were freed of meninges and gently triturated several times in culture medium using a flame-polished Pasteur pipette. Cells from one brain were plated in a 100-mm Petri dish and cultured in Dulbecco's modified Eagle's medium (DMEM)/F12 media (WelGene, Daegu, Korea) supplemented with N-2, B27 supplement (Gibco-Invitrogen, Carlsbad, CA, USA), 20 ng/ml epidermal growth factor (EGF), and basic fibroblast growth factor (bFGF; BD Biosciences, San Jose, CA, USA). EGF and bFGF were added every 2 days. For serial neurosphere formation, primary neurospheres were collected, incubated with Accumax (Millipore Corp, Bedford, MA, USA), and dissociated. For differentiation, dissociated cells were seeded on plates coated with 0.2 mg/ml poly-L-ornithine and 1 µg/ml fibronectin (Sigma, St. Louis, MO, USA) in the absence of growth factors or in the presence of CNTF (BD Biosciences).
Total RNA, including small RNA, was extracted using an miRNeasy Mini Kit isolation kit (Qiagen, Chatsworth, CA, USA) and reverse transcribed using a miScript Reverse Transcription kit (Qiagen) according to the manufacturers' instructions. miRNA levels were measured by Q-PCR using miScript SYBR Green PCR kit with miScript universal primer and miRNA-specific primers (Qiagen). RNU_6 was used as an endogenous control miRNA (Qiagen).
RNA (1000 ng) was prepared from 1-day-old mouse brains. Biotinylated RNA strands were hybridized at 48℃ for 18 hours on an Affymetrix GeneChip miRNA Array 4.0 (Affymetrix, Santa Clara, CA, US). The GeneChip was washed and stained in the Affymetrix Fluidics Station 450. Fluorescence signals amplified by the branched structure of a 3DNA dendrimer were scanned using an Affymetrix GeneChip Scanner 3000 7G.
For transfection, a mixture of Opti-MEM, 10 nM miRNA inhibitors or mimics, and RNAiMAX transfection reagent (Invitrogen, Carlsbad, CA, USA) was added to NSCs on differentiation day 1. Four days later, NSCs were prepared for experiments. Knockdown or overexpression of targets, including miR-326, miR-330 and miR-3099, was confirmed by Q-PCR. Possible cytotoxicity of miRNA inhibitors and mimics was analyzed using an LDH-Cytotoxicity Assay Kit (Biovision, Milpitas, CA, USA).
Cells were washed twice with cold phosphate-buffered saline (PBS) and lysed on ice with modified RIPA buffer (50 mM Tris-HCl pH 7.4, 1% NP-40, 0.25% Na-deoxycholate, 150 mM NaCl, 1 mM Na3VO4, 1 mM NaF) containing protease inhibitors (2 mM phenylmethylsulfonyl fluoride, 10 µg/ml leupeptin, 10 µg/ml pepstatin, 2 mM EDTA) and phosphatase inhibitor cocktail (GenDEPOT, Barker, TX, USA). Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to nitrocellulose membranes, and identified using antibodies specific for each protein. Antibodies for MAP2 were from Abcam (Cambridge, MA, USA); antibodies for TUJ-1 were from Covance (Berkeley, CA, USA); GFAP antibodies were from Sigma (St. Louis, MO, USA); and GAPDH antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Membranes were incubated with peroxidase-conjugated secondary antibodies (Jackson Immuno Research, West Grove, PA, USA) and visualized using an enhanced chemiluminescence system (Daeil Lab Inc., Seoul, Korea).
The effects of miRNAs were examined by measuring luciferase activity using a pGL3 control vector containing the 3'-UTR of GFAP, a pRL-
All data presented in this study are representative of at least three independent experiments. The statistical significance of differences between mean values of two groups was assessed by Student's t-test. For comparisons of more than two groups, we used a one-way analysis of variance (ANOVA) with Duncan's post hoc test.
In an effort to identify PINK1 functions in the brain, we analyzed expression of miRNAs in PINK1 wild-type (WT) and knockout (KO) brains. miRNAs were isolated from 1-day-old brains and analyzed using Affymetrix GeneChips. A PINK1 deficiency either increased or decreased several miRNAs; those that exhibited greater than a 2-fold change in expression in the absence of PINK1 are summarized in Table 1. Although the functions of most miRNAs whose expression was altered by PINK1 deficiency are not known, some of these miRNAs are known to be related to tumors and inflammation (Table 1).
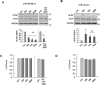
Previously, we found that PINK1 deficiency decreases protein but not mRNA levels of glial fibrillary acidic protein (GFAP), a marker of astrocytes, independently of protein degradation during brain development and differentiation of NSCs [39]. Thus, we examined expression levels of miRNAs that could regulate GFAP expression. We selected miR-326, miR-330, and miR-3099 since these three miRNAs are common candidates that bind to GFAP 3'-UTR and regulate translation based on data bases (TargetScan and miRanda). Using quantitative polymerase chain reaction (Q-PCR), we found that PINK1 deficiency decreased expression of all three miR-326, miR-330, and miR-3099. In the normal brain, expression levels of miR-326, miR-330, and miR-3099 gradually increased from embryonic day 12.5 (E12.5) to postnatal day 1~7 (P1~7), and slightly decreased (miR-3099) or further increased (miR-326 and 330) at 8 weeks (Fig. 1A). Interestingly, at P1, expression levels of all three miRNAs were lower in the PNIK1-KO brain than in the WT brain (Fig. 1B; miR-330 and miR-3099, p<0.05; miR-326, p=0.053).
Because expression of these miRNAs changed during development of the brain, we next examined expression of these miRNAs during differentiation of NSCs. After the induction of differentiation, the expression of markers of neurons (MAP2 and TUJ-1), astrocytes (GFAP), and oligodendrocytes (CNPase) increased at 2 days, and was further increased at 5 days (Fig. 2A). Interestingly, levels of all three miRNAs increased during differentiation of NSCs at 1 and 3 days (Fig. 2B). Furthermore, levels of these three miRNAs were significantly lower in PINK1-KO NSCs than in WT NSCs (Fig. 2C).
Because these miRNAs, particularly miR-326 and miR-330, are known to be related to astroglioma [29,40], we wondered whether these miRNAs regulated differentiation of NSCs into astrocytes. We first examined the expression levels of these miRNAs in the presence of CNTF, which drives differentiation of NSCs into astrocytes [41]. As expected, CNTF caused a concentration-dependent increase in GFAP expression (Fig. 2D). Under these differentiation-inducing conditions, expression of miR-326, miR-330 and miR-3099 increased (Fig. 2E); the levels of these miRNAs were also lower in KO NSCs than in WT NSCs (Fig. 2F).
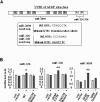
In serial experiments, we found that miR-326, miR-330, and miR-3099 all positively regulated GFAP expression. Specifically, inhibitors of these miRNAs significantly reduced GFAP levels in WT NSCs at differentiation day 5, without affecting TUJ-1 levels (Fig. 3A). Conversely, we found that mimics of miR-326, miR-330, and miR-3099 increased GFAP expression at this same time point in PINK1-KO NSCs, without changing expression of TUJ-1 (Fig. 3B). Neither inhibitors nor mimics of miRNAs affected cell viability (Fig. 3C, D).
Next, we examined whether these three miRNAs increase the translational efficacy of GFAP through direct binding to the 3'-UTR of GFAP mRNA using pGL3-luciferase constructs containing WT or GFAP 3'-UTR mutants because miRNAs regulate gene expression through binding to the 3'-UTR. GFAP 3'-UTR mutants were prepared by mutation of two common binding sites of miR-326 and miR-330 complementary sequences (CCCAGAG), or deletion of all eight miR-3099 binding sites (CTAGCCTA) (Fig. 4A). Consistent with expectations, application of miR-326, miR-330 or miR-3099 increased luciferase activity of normal GFAP 3'-UTR-containing constructs, but not that of GFAP constructs containing mutated 3'-UTRs (Fig. 4B). These results suggest that expression of miR-326, miR-330, and miR-3099 regulate GFAP expression during NSC differentiation. Taken together, our findings indicate that levels of the miRNAs, miR-326, miR-330 and miR-3099, may be regulated by PINK1 and contribute to the expression of GFAP during differentiation of NSCs.
In this study, we found that PINK1 deficiency changes expression of several miRNAs, including miR-326, miR-330 and miR-3099 that may be related to cell proliferation/cancer and inflammation (Table 1). We further found that miR-326, miR-330, and miR-3099 control GFAP expression during NSC differentiation, suggesting that a PINK1 deficiency could cause abnormal development of GFAP-expressing astrocytes.
Generally, miRNAs are thought to suppress mRNA translation. However, there are evidences to show that miRNAs can increase mRNA translation. For example, miR-369-3 levels are increased in HEK293T and Hela cells, and increase translation of the miR-369-3 target gene, tumor necrosis factor-α (TNF-α) [42]. Likewise, in
miRNAs are known to be linked to brain development and neurodegenerative diseases, including Alzheimer's disease, Huntington's disease, multiple sclerosis, and PD [25,26,27,28,29,30,31,32,33,34,35]. Notably, previous studies have reported possible link between neurodegenerative diseases and miR-326, miR-330, and miR-3099 [30,32,40,49,50]. miR-326 overexpression leads to severe multiple sclerosis [49] and regulates dopamine D2 receptor expression [30]. miR-330 enhances cell proliferation, promotes cell migration and invasion, and suppresses apoptosis by activating extracellular signal-regulated kinase (ERK) and phosphoinositide 3-kinase (PI3K)/AKT signaling pathways [40]. miR-330 is also associated with PD [32], and miR-3099 affects embryogenesis and neuronal differentiation [50]. In the current study, we discovered that miR-326, miR-330, and miR-3099 influence GFAP expression and/or astrogliogenesis. Furthermore, PINK1 was found to regulate expression of these miRNAs, suggesting a possible link between PINK1 and PD through these miRNAs.
For several decades, studies on PD and other neurodegenerative diseases have focused on neurons. However, genes related to neurodegenerative diseases are expressed in glia and NSCs, which means that mutations of these genes could cause defects in these cells. Therefore, it is also necessary to study neurodegenerative diseases in the context of these cells, which support neuronal survival and function in diverse ways.


- Valente EM, Bentivoglio AR, Dixon PH, Ferraris A, Ialongo T, Frontali M, Albanese A, Wood NW. Localization of a novel locus for autosomal recessive early-onset parkinsonism, PARK6, on human chromosome 1p35-p36. Am J Hum Genet 2001;68:895-900.
- Petit A, Kawarai T, Paitel E, Sanjo N, Maj M, Scheid M, Chen F, Gu Y, Hasegawa H, Salehi-Rad S, Wang L, Rogaeva E, Fraser P, Robinson B, St George-Hyslop P, Tandon A. Wild-type PINK1 prevents basal and induced neuronal apoptosis, a protective effect abrogated by Parkinson disease-related mutations. J Biol Chem 2005;280:34025-34032.
- Beilina A, Van Der Brug M, Ahmad R, Kesavapany S, Miller DW, Petsko GA, Cookson MR. Mutations in PTEN-induced putative kinase 1 associated with recessive parkinsonism have differential effects on protein stability. Proc Natl Acad Sci U S A 2005;102:5703-5708.
- Liu W, Vives-Bauza C, Acín-Peréz- R, Yamamoto A, Tan Y, Li Y, Magrané J, Stavarache MA, Shaffer S, Chang S, Kaplitt MG, Huang XY, Beal MF, Manfredi G, Li C. PINK1 defect causes mitochondrial dysfunction, proteasomal deficit and alpha-synuclein aggregation in cell culture models of Parkinson's disease. PLoS One 2009;4:e4597.
- Sim CH, Lio DS, Mok SS, Masters CL, Hill AF, Culvenor JG, Cheng HC. C-terminal truncation and Parkinson's disease-associated mutations down-regulate the protein serine/threonine kinase activity of PTEN-induced kinase-1. Hum Mol Genet 2006;15:3251-3262.
- Gandhi S, Wood-Kaczmar A, Yao Z, Plun-Favreau H, Deas E, Klupsch K, Downward J, Latchman DS, Tabrizi SJ, Wood NW, Duchen MR, Abramov AY. PINK1-associated Parkinson's disease is caused by neuronal vulnerability to calcium-induced cell death. Mol Cell 2009;33:627-638.
- Lin W, Kang UJ. Characterization of PINK1 processing, stability, and subcellular localization. J Neurochem 2008;106:464-474.
- Mills RD, Sim CH, Mok SS, Mulhern TD, Culvenor JG, Cheng HC. Biochemical aspects of the neuroprotective mechanism of PTEN-induced kinase-1 (PINK1). J Neurochem 2008;105:18-33.
- Valente EM, Abou-Sleiman PM, Caputo V, Muqit MM, Harvey K, Gispert S, Ali Z, Del Turco D, Bentivoglio AR, Healy DG, Albanese A, Nussbaum R, González-Maldonado R, Deller T, Salvi S, Cortelli P, Gilks WP, Latchman DS, Harvey RJ, Dallapiccola B, Auburger G, Wood NW. Hereditary early-onset Parkinson's disease caused by mutations in PINK1. Science 2004;304:1158-1160.
- Zhou C, Huang Y, Shao Y, May J, Prou D, Perier C, Dauer W, Schon EA, Przedborski S. The kinase domain of mitochondrial PINK1 faces the cytoplasm. Proc Natl Acad Sci U S A 2008;105:12022-12027.
- Choi I, Kim J, Jeong HK, Kim B, Jou I, Park SM, Chen L, Kang UJ, Zhuang X, Joe EH. PINK1 deficiency attenuates astrocyte proliferation through mitochondrial dysfunction, reduced AKT and increased p38 MAPK activation, and downregulation of EGFR. Glia 2013;61:800-812.
- Deng H, Jankovic J, Guo Y, Xie W, Le W. Small interfering RNA targeting the PINK1 induces apoptosis in dopaminergic cells SH-SY5Y. Biochem Biophys Res Commun 2005;337:1133-1138.
- Haque ME, Thomas KJ, D'Souza C, Callaghan S, Kitada T, Slack RS, Fraser P, Cookson MR, Tandon A, Park DS. Cytoplasmic Pink1 activity protects neurons from dopaminergic neurotoxin MPTP. Proc Natl Acad Sci U S A 2008;105:1716-1721.
- Kim J, Byun JW, Choi I, Kim B, Jeong HK, Jou I, Joe E. PINK1 deficiency enhances inflammatory cytokine release from acutely prepared brain slices. Exp Neurobiol 2013;22:38-44.
- Lin W, Wadlington NL, Chen L, Zhuang X, Brorson JR, Kang UJ. Loss of PINK1 attenuates HIF-1α induction by preventing 4E-BP1-dependent switch in protein translation under hypoxia. J Neurosci 2014;34:3079-3089.
- Requejo-Aguilar R, Lopez-Fabuel I, Fernandez E, Martins LM, Almeida A, Bolaños JP. PINK1 deficiency sustains cell proliferation by reprogramming glucose metabolism through HIF1. Nat Commun 2014;5:4514.
- Miska EA, Alvarez-Saavedra E, Townsend M, Yoshii A, Sestan N, Rakic P, Constantine-Paton M, Horvitz HR. Microarray analysis of microRNA expression in the developing mammalian brain. Genome Biol 2004;5:R68.
- Moreau MP, Bruse SE, Jornsten R, Liu Y, Brzustowicz LM. Chronological changes in microRNA expression in the developing human brain. PLoS One 2013;8:e60480.
- Ziats MN, Rennert OM. Identification of differentially expressed microRNAs across the developing human brain. Mol Psychiatry 2014;19:848-852.
- Fineberg SK, Kosik KS, Davidson BL. MicroRNAs potentiate neural development. Neuron 2009;64:303-309.
- Zheng K, Li H, Huang H, Qiu M. MicroRNAs and glial cell development. Neuroscientist 2012;18:114-118.
- Eacker SM, Dawson TM, Dawson VL. Understanding microRNAs in neurodegeneration. Nat Rev Neurosci 2009;10:837-841.
- Hébert SS, De Strooper B. Alterations of the microRNA network cause neurodegenerative disease. Trends Neurosci 2009;32:199-206.
- Meza-Sosa KF, Valle-García D, Pedraza-Alva G, Pérez-Martínez L. Role of microRNAs in central nervous system development and pathology. J Neurosci Res 2012;90:1-12.
- Boissonneault V, Plante I, Rivest S, Provost P. MicroRNA-298 and microRNA-328 regulate expression of mouse beta-amyloid precursor protein-converting enzyme 1. J Biol Chem 2009;284:1971-1981.
- Wang WX, Rajeev BW, Stromberg AJ, Ren N, Tang G, Huang Q, Rigoutsos I, Nelson PT. The expression of microRNA miR-107 decreases early in Alzheimer's disease and may accelerate disease progression through regulation of betasite amyloid precursor protein-cleaving enzyme 1. J Neurosci 2008;28:1213-1223.
- Packer AN, Xing Y, Harper SQ, Jones L, Davidson BL. The bifunctional microRNA miR-9/miR-9* regulates REST and CoREST and is downregulated in Huntington's disease. J Neurosci 2008;28:14341-14346.
- Ma X, Zhou J, Zhong Y, Jiang L, Mu P, Li Y, Singh N, Nagarkatti M, Nagarkatti P. Expression, regulation and function of microRNAs in multiple sclerosis. Int J Med Sci 2014;11:810-818.
- Kim Y, Kim H, Park H, Park D, Lee H, Lee YS, Choe J, Kim YM, Jeoung D. miR-326-histone deacetylase-3 feedback loop regulates the invasion and tumorigenic and angiogenic response to anti-cancer drugs. J Biol Chem 2014;289:28019-28039.
- Shi S, Leites C, He D, Schwartz D, Moy W, Shi J, Duan J. MicroRNA-9 and microRNA-326 regulate human dopamine D2 receptor expression, and the microRNA-mediated expression regulation is altered by a genetic variant. J Biol Chem 2014;289:13434-13444.
- Gehrke S, Imai Y, Sokol N, Lu B. Pathogenic LRRK2 negatively regulates microRNA-mediated translational repression. Nature 2010;466:637-641.
- Chandrasekaran S, Bonchev D. A network view on Parkinson's disease. Comput Struct Biotechnol J 2013;7:e201304004.
- Doxakis E. Post-transcriptional regulation of alpha-synuclein expression by mir-7 and mir-153. J Biol Chem 2010;285:12726-12734.
- Farh KK, Grimson A, Jan C, Lewis BP, Johnston WK, Lim LP, Burge CB, Bartel DP. The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science 2005;310:1817-1821.
- Junn E, Lee KW, Jeong BS, Chan TW, Im JY, Mouradian MM. Repression of alpha-synuclein expression and toxicity by microRNA-7. Proc Natl Acad Sci U S A 2009;106:13052-13057.
- Kim JH, Jou I, Joe EH. Suppression of miR-155 expression in IFN-γ-treated astrocytes and microglia by DJ-1: a possible mechanism for maintaining SOCS1 expression. Exp Neurobiol 2014;23:148-154.
- Xiong H, Wang D, Chen L, Choo YS, Ma H, Tang C, Xia K, Jiang W, Ronai Z, Zhuang X, Zhang Z. Parkin, PINK1, and DJ-1 form a ubiquitin E3 ligase complex promoting unfolded protein degradation. J Clin Invest 2009;119:650-660.
- Kim YH, Chung JI, Woo HG, Jung YS, Lee SH, Moon CH, Suh-Kim H, Baik EJ. Differential regulation of proliferation and differentiation in neural precursor cells by the Jak pathway. Stem Cells 2010;28:1816-1828.
- Choi I, Choi DJ, Yang H, Woo JH, Chang MY, Kim JY, Sun W, Park SM, Jou I, Lee SH, Joe EH. PINK1 expression increases during brain development and stem cell differentiation, and affects the development of GFAP-positive astrocytes. Mol Brain 2016;9:5.
- Yao Y, Xue Y, Ma J, Shang C, Wang P, Liu L, Liu W, Li Z, Qu S, Li Z, Liu Y. MiR-330-mediated regulation of SH3GL2 expression enhances malignant behaviors of glioblastoma stem cells by activating ERK and PI3K/AKT signaling pathways. PLoS One 2014;9:e95060.
- Setoguchi T, Kondo T. Nuclear export of OLIG2 in neural stem cells is essential for ciliary neurotrophic factor-induced astrocyte differentiation. J Cell Biol 2004;166:963-968.
- Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science 2007;318:1931-1934.
- Mortensen RD, Serra M, Steitz JA, Vasudevan S. Posttranscriptional activation of gene expression in Xenopus laevis oocytes by microRNA-protein complexes (microRNPs). Proc Natl Acad Sci U S A 2011;108:8281-8286.
- Truesdell SS, Mortensen RD, Seo M, Schroeder JC, Lee JH, LeTonqueze O, Vasudevan S. MicroRNA-mediated mRNA translation activation in quiescent cells and oocytes involves recruitment of a nuclear microRNP. Sci Rep 2012;2:842.
- Lin CC, Liu LZ, Addison JB, Wonderlin WF, Ivanov AV, Ruppert JM. A KLF4-miRNA-206 autoregulatory feedback loop can promote or inhibit protein translation depending upon cell context. Mol Cell Biol 2011;31:2513-2527.
- Henke JI, Goergen D, Zheng J, Song Y, Schüttler CG, Fehr C, Jünemann C, Niepmann M. microRNA-122 stimulates translation of hepatitis C virus RNA. EMBO J 2008;27:3300-3310.
- Saraiya AA, Li W, Wang CC. Transition of a microRNA from repressing to activating translation depending on the extent of base pairing with the target. PLoS One 2013;8:e55672.
- Vasudevan S. Posttranscriptional upregulation by microRNAs. Wiley Interdiscip Rev RNA 2012;3:311-330.
- Du C, Liu C, Kang J, Zhao G, Ye Z, Huang S, Li Z, Wu Z, Pei G. MicroRNA miR-326 regulates TH-17 differentiation and is associated with the pathogenesis of multiple sclerosis. Nat Immunol 2009;10:1252-1259.
- Ling KH, Brautigan PJ, Hahn CN, Daish T, Rayner JR, Cheah PS, Raison JM, Piltz S, Mann JR, Mattiske DM, Thomas PQ, Adelson DL, Scott HS. Deep sequencing analysis of the developing mouse brain reveals a novel microRNA. BMC Genomics 2011;12:176.



