Articles
Article Tools
Stats or Metrics
Article
Original Article
Exp Neurobiol 2017; 26(6): 350-361
Published online December 31, 2017
https://doi.org/10.5607/en.2017.26.6.350
© The Korean Society for Brain and Neural Sciences
AAV-Mediated Astrocyte-Specific Gene Expression under Human ALDH1L1 Promoter in Mouse Thalamus
Wuhyun Koh1,2, Yongmin Mason Park1,2, Seung Eun Lee3* and C. Justin Lee1,2,4*
1Division of Bio-Medical Science &Technology, Department of Neuroscience, KIST School, Korea University of Science and Technology, Seoul 02792, 2Center for Neuroscience and Functional Connectomics, Korea Institute of Science and Technology (KIST), Seoul 02792, 3Virus Facility, Research Animal Resource Center, Korea Institute of Science and Technology (KIST), Seoul 02792, 4Center for Glia-Neuron Interaction, Korea Institute of Science and Technology (KIST), Seoul 02792, Korea
Correspondence to: *To whom correspondence should be addressed.
C. Justin Lee, TEL: 82-2-958-6940, FAX: 82-2-958-6937
e-mail: cjl@kist.re.kr
Seung Eun Lee, TEL: 82-2-958-6959, FAX: 82-2-958-6937
e-mail: selee@kist.re.kr
Abstract
Adeno-associated virus (AAV)-mediated gene delivery has been proposed to be an essential tool of gene therapy for various brain diseases. Among several cell types in the brain, astrocyte has become a promising therapeutic target for brain diseases, as more and more contribution of astrocytes in pathophysiology has been revealed. Until now, genetically targeting astrocytes has been possible by utilizing the
Graphical Abstract
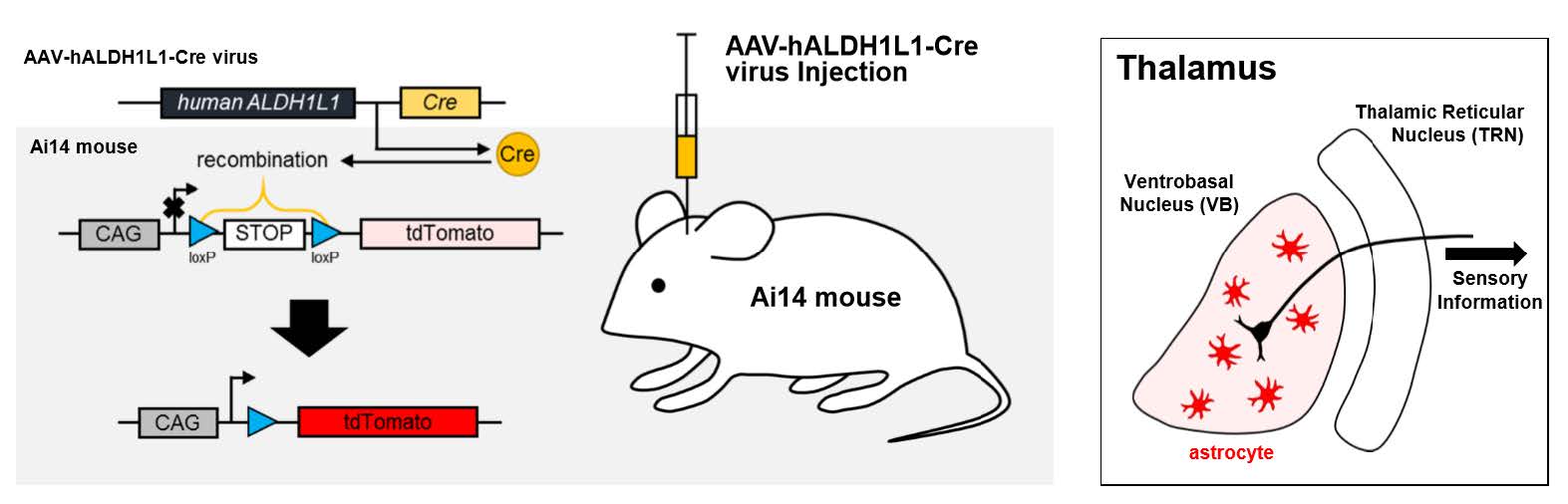
Keywords: Astrocytes, Dependovirus, Ventral Thalamic Nuclei, ALDH1L1 protein, human, Cre recombinase, Glial Fibrillary
Acidic Protein
INTRODUCTION
In the central nervous system, the most abundant cell type is astrocyte, which has been revealed to have a number of functions, including modulation of synaptic transmission [1,2], relay for nutrients to adjacent neuron [3], clearance of waste molecules [4]. To study functions of astrocyte in many brain regions, researchers have been relying heavily on the conventional astrocyte-specific marker, glial fibrillary acidic protein (GFAP) and its promoter after the discovery of its specificity for astrocytes [5]. However, it also has been reported that GFAP expression is low in several brain areas, including thalamus [6]. GFAP also has been known to be expressed transiently in neural progenitor cells, which could lead to neuronal expression near neurogenic areas [7]. Therefore, utilizing GFAP may not be optimal in those brain areas. Moreover, there is a pressing need for an alternative astrocyte-specific marker to study astrocytic functions in those brain areas.
There are two superb examples of utilizing astrocyte-specific promoter to study and target astrocytes in the mouse brain. First one is the generation of transgenic mouse lines such as GFAP-EGFP mouse [8] and GFAP-Cre/ERT2 mouse [9]. Although utilizing transgenic mouse is one of the best choices to target astrocytes, it could be very resource- and time-consuming when they need to be transferred or maintained. It is also difficult to extend the study to other species beyond mouse. Another example is to apply astrocyte specific promoter in virus applications such as in GFAP-GFP virus [10]. Virus containing astrocyte-specific promoter allows researchers to examine the effects of a gene of interest in local astrocytes in a specific brain area where the virus is injected. Moreover, virus is easy to apply to different species, such as mouse, marmoset, monkey or even human. As more and more contribution of astrocytes in pathophysiology of brain diseases has been revealed, astrocyte-specific targeting with virus would be critically essential as a potential therapeutic tool [11]. Among viruses, adeno-associated virus (AAV) has become very popular with many prominent advantages: AAV is able to infect both dividing and non-dividing cell types with less immunogenic effect [12]. Ultimately, AAV promises to be the best available option for gene therapy for brain diseases [13,14].
As an alternative to GFAP promoter, aldehyde dehydrogenase family 1, member L1 (ALDH1L1) was recently identified as a new astrocyte-specific marker in the brain [15]. The promoter of ALDH1L1 should be useful in brain regions where GFAP expression is low. There has been attempts to characterize the
In this study, we constructed and characterized human
MATERIALS AND METHODS
The h
The viral vectors were pseudotyped, where the transgene of interest was flanked by inverted terminal repeats of the AAV2 packaged in an AAV-DJ capsid. AAV-DJ was engineered via DNA family shuffling technology which created a hybrid capsid from 8 AAV serotype. AAV-hALDH1L1-Cre vector was thereafter purified by iodixanol gradients by the KIST Virus Facility (http://virus.kist.re.kr). The production titer was 1.5×1013 genome copies/ml (GC/ml).
Adult (aged 8~10 weeks) male and female Ai14 mice (RCL-tdTomato; Rosa-CAG-LSL-tdTomato-WPRE::deltaNeo) in C57BL/6J strain were used as transgenic reporter line. Mice had free access to food and water and were kept on a 12 hours light-dark cycle. All experimental procedures described below were performed in accordance with the Institutional guidelines for experimental animal care and use of the Korea Institute of Science and Technology (KIST; Seoul, Korea).
Mice were anesthetized with 2% avertin (20 µl/g, i.p.) and placed on stereotaxic apparatus (Kopf instrument, USA). AAV-hALDH1L1-Cre virus was injected through 33 gauge blunt NanoFil needle (World Precision Instruments, USA), connected with 10 µl Hamilton micro syringe (Hamilton, Switzerland) filled with distilled water. Syringe pump (KD Scientific, USA) was used to inject the virus with the rate of 0.2 µl per minute. Needle was gently placed into target regions (thalamic VB: -1.8 mm AP, ±1.6 mm ML, 3.75 mm (1 µl) and 3.25 mm (1 µl) from brain surface; hippocampal CA1: -1.8 mm AP, ±1.6 mm ML, 1.6 mm (2 µl) from brain surface; BLA: -1.8 mm AP, ±3.4 mm ML, 4.55 mm (2 µl) from brain surface; mPFC: 1.8 mm AP, ±0.3 mm ML, 1.7 mm (2 µl) from brain surface), bilaterally. Mice were sacrificed after 10 days recovery.
Mice were anesthetized with 2% avertin (20 µl/g, i.p.) and perfused with 0.1 M phosphate buffered saline (PBS) at room temperature followed by ice-cold 4% paraformaldehyde. Extracted brains were post-fixed in 4% paraformaldehyde at 4℃ overnight. Post-fixed brains were incubated in 30% sucrose at 4℃ for more than 24 hours. Coronal brain sections were prepared with 30 µm thickness in cryostat microtome (Thermo Scientific). Brain sections were rinsed three times with PBS, and blocking step was performed with a blocking solution (0.3% Triton X-100 (Sigma), 2% donkey serum (Millipore) and 2% goat serum (Abcam) in 0.1 M PBS) for 1.5 hours. Blocking step was followed by application of a mixture of primary antibodies as follows, rabbit anti-Aldh1L1 antibody (Novus), mouse anti-NeuN antibody (Millipore). Sections were incubated overnight at 4℃ with shaking for primary antibodies. After overnight incubation, the sections were washed three times in PBS and then incubated in corresponding fluorescent secondary antibodies for 1.5 hours. In three times wash process of secondary antibodies, they also were counterstained with 4′,6′-diamidino-2-phenylindole (DAPI, Pierce) before they were mounted onto slide glass (Thermo Scientific). A series of fluorescence images were obtained with Nikon A1 confocal microscope, and 30 µm Z stack images in 3-µm steps were processed for further analysis using NIS-Elements (Nikon) software and ImageJ (NIH) program. Reference atlases of mouse brain in the figure were obtained from Allen Brain Atlas (Reference Atlas, Version 2 (2011)) [21].
Primary cortical astrocytes were cultured from P0 to P3 C57BL/6N mouse pups. The cerebral cortex was dissected free meninges and softly triturating to single cell unit. Culture media was prepared as follow. Dulbecco's modified Eagle's medium (DMEM, Invitrogen) supplemented with 25 mM glucose, 2 mM glutamine, 10% heat-inactivated horse serum, 10% heat-inactivated fetal bovine serum and 1,000 units/ml penicillin-streptomycin. On the third day after culture, cells were rinsed with repeated gentle pipetting and the media was replaced to get rid of debris. The day before treatment of viruses, cells were transferred to coverslip (1×104 per coverslip) coated with 0.1 mg/ml Poly D-Lysine (PDL, Sigma). Cultured cells were maintained at 37℃ in a humidified 5% CO2 incubator.
Three days after virus transduction, cultured astrocytes on coverslip were rinsed with PBS and fixed with 4% paraformaldehyde for 20 minutes. After three times wash with PBS, immunocytochemistry was performed as similar manner in IHC as describe above. Briefly, blocking step was performed with a blocking solution (same with IHC, but no goat serum) for 1 hour. Blocking step was followed by application of primary antibody, rabbit anti-Aldh1L1 antibody for 2 hours, RT. After that, the cells were washed in PBS and then incubated in corresponding fluorescent secondary antibodies for 1 hour. In three times wash process of secondary antibodies, they also were counterstained with DAPI before coverslips were mounted onto slide glass.
RESULTS
Aldh1L1 has been shown to be a highly specific marker for astrocytes with a substantially broader pattern of astrocytic expression than the conventional astrocyte marker, GFAP [14]. We sought to develop h
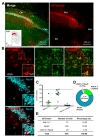
To test cell-type specific expression of Cre recombinase by h
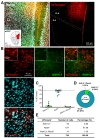
Next, we tested astrocyte-specific Cre expression in amygdala and hippocampus. AAV-hALDH1L1-Cre virus was delivered to BLA and hippocampal CA1, respectively (Fig. 3A and 4A). In BLA, both cell types, astrocytes and neurons, showed tdTomato fluorescence (Fig. 3B). Average percentage of Aldh1L1+/tdTomato+ cells in each image was lower than that of NeuN+/tdTomato+ cells (Aldh1L1+: 26.5±7.5% versus NeuN+: 69.5±8.7%) (Fig. 3C). Among the analyzed total Aldh1L1+ cells, tdTomato+ cells were 28.35%, and NeuN+ cells were 68.13% (Fig. 3D and E). These results indicate that AAV-hALDH1L1-Cre virus expresses Cre recombinase in both astrocytes and neurons in BLA.
Similar to BLA, in hippocampal CA1, AAV-hALDH1L1-Cre virus induced tdTomato fluorescence from astrocytes and neurons in Ai14 mouse (Fig. 4B). Average percentage of Aldh1L1+/tdTomato+ cells in each image was 20.4±8.8%, which is lower than that of NeuN+/tdTomato+ cells (75.3±8.6%) (Fig. 4C). In total of analyzed tdTomato+ cells in hippocampal CA1 area, a population of Aldh1L1+ cells was 18.00%, and a population of NeuN+ cells was 76.25% (Fig. 4D and E). These results indicate that AAV-hALDH1L1-Cre virus expresses Cre recombinase in both astrocytes and neurons in hippocampal CA1.
We further investigated activity pattern of h
Taken together, expression pattern of AAV-hALDH1L1-Cre virus-mediated tdTomato fluorescence was different in different brain areas (Table 1). Additionally, we further analyzed a proportion of astrocytes that express tdTomato in these brain regions. To measure the proportion of astrocytes expressing tdTomato fluorescence, we counted all Aldh1L1+ cells near the infected areas from each brain region, and the number of tdTomato+ cells with Aldh1L1+ was divided by the total number of Aldh1L1+ cells. Calculated proportions of astrocytes expressing tdTomato were as following, VB: 81.91%; BLA: 51.03%; CA1: 25.55%; mPFC: 13.26%. These results indicate that astrocytes in VB, BLA, CA1 and mPFC are heterogeneous in virus transduction efficacy or h
As a summary, AAV-hALDH1L1-Cre virus showed highly specific expression in astrocyte of thalamic area. The virus-mediated tdTomato fluorescence could be observed in both astrocyte and neuron in amygdala and hippocampus. Finally, AAV-hALDH1L1-Cre virus in mPFC showed highly specific expression in neuron.
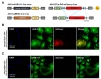
We further investigated the efficiency of AAV-hALDH1L1-Cre virus in cultured system. To examine the Cre recombinase expression under h
DISCUSSION
Our study is the first development and characterization of h
One possible reason is that serotype of AAV virus could affect cell-type specificity of virus infection, as many studies previously reported [22,23,24]. Natural AAV viruses have a specific pattern of infection that reflects the interaction and recognition between the viral capsid, envelope and receptors expressed in susceptible cells. In particular, although AAV serotype 4 and AAV rh43 are known to target astrocytes [7,25], the receptor of this serotype has not yet been identified. Moreover, pseudotype-dependent lentivirus has been utilized to target for astrocyte. In the current study, we utilized AAV-DJ serotype to target astrocytes with h
Another possible reason could be that h
Lastly, the neurons that expressed tdTomato fluorescence by h
In conclusion, AAV-mediated gene expression under h
Figures
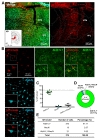
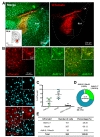
Tables
| Cell types | Thalamic ventrobasal nucleus (VB) | Basolateral amygdala (BLA) | Hippocampal CA1 | Medial prefrontal cortex (mPFC) |
|---|---|---|---|---|
| Astrocytes (%) | 91.71 | 28.35 | 18.00 | 6.66 |
| Neurons (%) | 2.67 | 68.13 | 76.25 | 91.98 |
| Others (%) | 5.61 | 3.52 | 5.75 | 1.36 |
| Astrocytic transduction efficiency (%) | 81.91 | 51.03 | 25.55 | 13.26 |
References
- Haydon PG, Carmignoto G. Astrocyte control of synaptic transmission and neurovascular coupling. Physiol Rev 2006;86:1009-1031.
- Jo S, Yarishkin O, Hwang YJ, Chun YE, Park M, Woo DH, Bae JY, Kim T, Lee J, Chun H, Park HJ, Lee DY, Hong J, Kim HY, Oh SJ, Park SJ, Lee H, Yoon BE, Kim Y, Jeong Y, Shim I, Bae YC, Cho J, Kowall NW, Ryu H, Hwang E, Kim D, Lee CJ. GABA from reactive astrocytes impairs memory in mouse models of Alzheimer's disease. Nat Med 2014;20:886-896.
- Stobart JL, Anderson CM. Multifunctional role of astrocytes as gatekeepers of neuronal energy supply. Front Cell Neurosci 2013;7:38.
- Xie L, Kang H, Xu Q, Chen MJ, Liao Y, Thiyagarajan M, O'donnell J, Christensen DJ, Nicholson C, Iliff JJ, Takano T, Deane R, Nedergaard M. Sleep drives metabolite clearance from the adult brain. Science 2013;342:373-377.
- Jacque CM, Vinner C, Kujas M, Raoul M, Racadot J, Baumann NA. Determination of glial fibrillary acidic protein (GFAP) in human brain tumors. J Neurol Sci 1978;35:147-155.
- Kálmán M, Hajós F. Distribution of glial fibrillary acidic protein (GFAP)-immunoreactive astrocytes in the rat brain. I. Forebrain. Exp Brain Res 1989;78:147-163.
- Liu Y, Namba T, Liu J, Suzuki R, Shioda S, Seki T. Glial fibrillary acidic protein-expressing neural progenitors give rise to immature neurons via early intermediate progenitors expressing both glial fibrillary acidic protein and neuronal markers in the adult hippocampus. Neuroscience 2010;166:241-251.
- Nolte C, Matyash M, Pivneva T, Schipke CG, Ohlemeyer C, Hanisch UK, Kirchhoff F, Kettenmann H. GFAP promoter-controlled EGFP-expressing transgenic mice: a tool to visualize astrocytes and astrogliosis in living brain tissue. Glia 2001;33:72-86.
- Hirrlinger PG, Scheller A, Braun C, Hirrlinger J, Kirchhoff F. Temporal control of gene recombination in astrocytes by transgenic expression of the tamoxifen-inducible DNA recombinase variant CreERT2. Glia 2006;54:11-20.
- Shinohara Y, Konno A, Takahashi N, Matsuzaki Y, Kishi S, Hirai H. Viral vector-based dissection of marmoset GFAP promoter in mouse and marmoset brains. PLoS One 2016;11:e0162023.
- Almad AA, Maragakis NJ. Glia: an emerging target for neurological disease therapy. Stem Cell Res Ther 2012;3:37.
- Lakhan R, Baylink DJ, Lau KH, Tang X, Sheng MH, Rundle CH, Qin X. Local administration of AAV-DJ pseudoserotype expressing COX2 provided early onset of transgene expression and promoted bone fracture healing in mice. Gene Ther 2015;22:721-728.
- Daya S, Berns KI. Gene therapy using adeno-associated virus vectors. Clin Microbiol Rev 2008;21:583-593.
- Ortolano S, Spuch C, Navarro C. Present and future of adeno associated virus based gene therapy approaches. Recent Pat Endocr Metab Immune Drug Discov 2012;6:47-66.
- Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christopherson KS, Xing Y, Lubischer JL, Krieg PA, Krupenko SA, Thompson WJ, Barres BA. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci 2008;28:264-278.
- Heintz N. Gene expression nervous system atlas (GENSAT). Nat Neurosci 2004;7:483.
- Srinivasan R, Lu TY, Chai H, Xu J, Huang BS, Golshani P, Coppola G, Khakh BS. New transgenic mouse lines for selectively targeting astrocytes and studying calcium signals in astrocyte processes in situ and in vivo. Neuron 2016;92:1181-1195.
- Mudannayake JM, Mouravlev A, Fong DM, Young D. Transcriptional activity of novel ALDH1L1 promoters in the rat brain following AAV vector-mediated gene transfer. Mol Ther Methods Clin Dev 2016;3:16075.
- Thoms JA, Birger Y, Foster S, Knezevic K, Kirschenbaum Y, Chandrakanthan V, Jonquieres G, Spensberger D, Wong JW, Oram SH, Kinston SJ, Groner Y, Lock R, MacKenzie KL, Göttgens B, Izraeli S, Pimanda JE. ERG promotes T-acute lymphoblastic leukemia and is transcriptionally regulated in leukemic cells by a stem cell enhancer. Blood 2011;117:7079-7089.
- Park J, Throop AL, LaBaer J. Site-specific recombinational cloning using gateway and in-fusion cloning schemes. Curr Protoc Mol Biol 2015;110:3.20.1-3.20.23.
- Dong HW. The Allen reference atlas: a digital color brain atlas of the C57Bl/6J male mouse. Wiley: Chichester, (2008).
- Merienne N, Le Douce J, Faivre E, Déglon N, Bonvento G. Efficient gene delivery and selective transduction of astrocytes in the mammalian brain using viral vectors. Front Cell Neurosci 2013;7:106.
- Pignataro D, Sucunza D, Vanrell L, Lopez-Franco E, Dopeso-Reyes IG, Vales A, Hommel M, Rico AJ, Lanciego JL, Gonzalez-Aseguinolaza G. Adeno-associated viral vectors serotype 8 for cell-specific delivery of therapeutic genes in the central nervous system. Front Neuroanat 2017;11:2.
- Royo NC, Vandenberghe LH, Ma JY, Hauspurg A, Yu L, Maronski M, Johnston J, Dichter MA, Wilson JM, Watson DJ. Specific AAV serotypes stably transduce primary hippocampal and cortical cultures with high efficiency and low toxicity. Brain Res 2008;1190:15-22.
- Lawlor PA, Bland RJ, Mouravlev A, Young D, During MJ. Efficient gene delivery and selective transduction of glial cells in the mammalian brain by AAV serotypes isolated from nonhuman primates. Mol Ther 2009;17:1692-1702.
- Oleinik NV, Krupenko NI, Krupenko SA. Epigenetic silencing of ALDH1L1, a metabolic regulator of cellular proliferation, in cancers. Genes Cancer 2011;2:130-139.
- Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev 2011;25:1010-1022.



