Articles
Article Tools
Stats or Metrics
Article
Original Article
Exp Neurobiol 2019; 28(2): 229-246
Published online April 30, 2019
https://doi.org/10.5607/en.2019.28.2.229
© The Korean Society for Brain and Neural Sciences
Autophagy Mediates Astrogenesis in Adult Hippocampal Neural Stem Cells
Shinwon Ha1,†, Seol-Hwa Jeong1,†, Kyungrim Yi1,†, Jamie Jeong-Min Chu1, Seolsong Kim1, Eun-Kyoung Kim1,2, and Seong-Woon Yu1,2*
1Department of Brain and Cognitive Sciences, Daegu Gyeongbuk Institute of Science and Technology (DGIST), Daegu 42988, Korea.
2Neurometabolomics Research Center, Daegu Gyeongbuk Institute of Science and Technology (DGIST), Daegu 42988, Korea.
Correspondence to: *To whom correspondence should be addressed.
TEL: 82-53-785-6113, FAX: 82-53-785-1209
e-mail: yusw@dgist.ac.kr
†These authors contributed equally to this work.
Abstract
Neural stem cells (NSCs) have the ability to self-renew and differentiate into neurons, oligodendrocytes, and astrocytes. Highly dynamic nature of NSC differentiation requires the intimate involvement of catabolic processes such as autophagy. Autophagy is a major intracellular degradation pathway necessary for cellular homeostasis and remodeling. Autophagy is important for mammalian development and its role in neurogenesis has recently drawn much attention. However, little is known about how autophagy is associated with differentiation of NSCs into other neural lineages. Here, we report that autophagy plays a critical role in differentiation of adult rat hippocampal neural stem (HCN) cells into astrocytes. During differentiation, autophagy flux peaked at early time points, and remained high. Pharmacological or genetic suppression of autophagy by stable knockdown of Atg7, LC3 or CRISPR-Cas9-mediated knockout (KO) of p62 impaired astrogenesis, while reintroduction of p62 recovered astrogenesis in p62 KO HCN cells. Taken together, our findings suggest that autophagy plays a key role in astrogenesis in adult NSCs.
Graphical Abstract
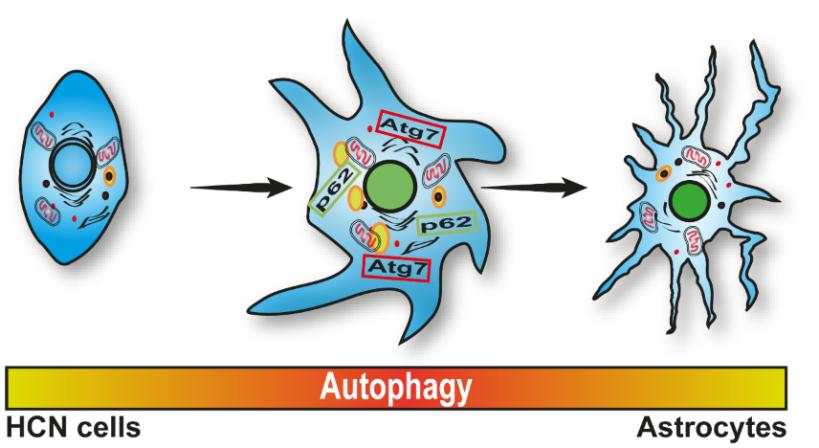
Keywords: Adult stem cells, Astrocytes, Autophagy, Autophagy-related protein 7, Cell differentiation, Neural stem cells, Sequestosome-1 protein
INTRODUCTION
Autophagy is an evolutionarily conserved, major intracellular degradation pathway [1]. Autophagy is initiated by phagophore formation and its expansion to generate autophagosomes [2,3]. Upon completion of autophagosome formation with cargos inside, autophagosome fuses with a lysosome to become autolysosome, where its contents are degraded by acidic proteases [4]. Subsequently, degraded products are transported back to the cytoplasm to be reused as a source of energy [5]. There are several methods to monitor autophagy. Microtubule-associated protein light chain 3 (LC3, a mammalian homologue of yeast Atg8) is the most widely used autophagy-related protein used to measure autophagosome formation [6]. LC3 is a ubiquitin-like protein that is, upon synthesis, cleaved by Atg4 protease to generate the cytosolic form [7]. Then, at sites of autophagosome formation, LC3 is conjugated to phosphatidylethanolamine through the action of the E1-like activating enzyme Atg7 and the E2-like conjugating enzyme Atg3 [3,7,8]. The non-conjugated, cytosolic form is referred to as LC3-I and the phosphatidylethanolamine-conjugated, autophagic form as LC3-II [3]. An increase in LC3-II level as a marker of autophagosome formation can be measured by Western blotting for autophagy examination [9]. The appearance of puncta- or dot-like signals of a fluorescent-tagged LC3 protein is also indicative of autophagosome formation [6,10]. p62 is an autophagy adaptor protein regulating the packaging and delivery of organelles and polyubiquitinated, misfolded proteins into autophagosomes for clearance [11]. During autophagic degradation, p62 is also degraded with the substrates, so a decrease in p62 protein amount is regarded as another marker of autophagy induction [11,12,13].
Increased autophagy flux includes not only more autophagosome formation, but also efficient fusion of autophagosomes and lysosomes for delivery of autophagic substrate cargos and their degradation [14]. Therefore, blocking the late autophagy steps, such as fusion between autophagosomes and lysosomes, with bafilomycin A1 (Baf.A1) can be used to distinguish whether an increase in autophagy markers is due to genuine up-regulation of autophagy flux (on-rate) or impaired autophagy flux (off-rate) [5,15]. Blocking of autophagosome fusion under high autophagy flux conditions is expected to increase accumulation of LC3-II and p62, while an increase in autophagy markers such as LC3-II or LC3 puncta resulting from impaired autophagy will not be affected by additional blocking of autophagy with Baf.A1 [6,15]. Since the pH inside autophagosomes differs from that inside autolysosomes, several assay methods based on pH have been developed to assess autophagy flux, including LC3 tagged with two fluorescent proteins, monomeric RFP and GFP, arranged in tandem (mRFP-GFP-LC3). Monitoring autophagy flux with mRFP-GFP-LC3 is based on different pH stability of mRFP and GFP proteins. The presence of GFP and RFP in autophagosomes gives yellow fluorescent signals; however, since GFP is easily quenched under acidic conditions, such as those in lysosomes, only RFP signals are retained after fusion of autophagosomes with lysosomes. Therefore, an increase in both the total number of puncta and red-to-yellow signal ratio indicates an increase in autophagy flux [16].
Since autophagy is readily inducible in response to various environmental cues and cellular stressors, autophagy is highly active during differentiation and development [17]. The critical role of autophagy in mammalian development has been well documented in mice lacking the Atg genes including
To address the role of autophagy in astrocyte differentiation, we used murine adult NSCs, which were derived from the hippocampus of a 2-month-old male rat. The discovery of persistent generation of new neurons in the brain throughout adulthood has sparked interest in the role of adult neurogenesis in brain function and plasticity [30,31]. Hippocampus is one of the two well-established regions of adult neurogenesis [32]. Since hippocampus is deeply involved in learning and memory, mood regulation, reward-seeking and other neurobehaviors, understanding the pathophysiological roles of adult hippocampal neurogenesis and its underlying neural mechanisms are of great interest [33,34]. Various
MATERIALS AND METHODS
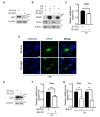
Baf.A1 (BML-CM110, Enzo, USA), pepstatin A (PepA, P5318, Sigma-Aldrich, USA), E64d (E8640, Sigma-Aldrich), puromycin (NC9138068, Invitrogen, USA), hygromycin B (H0192, Duchefa, The Netherlands), fetal bovine serum (FBS, #101, Tissue Culture Biologicals, USA), forskolin (BML-CN100, Enzo), and retinoic acid (RA, #BML-GR100, Enzo) were purchased from the indicated companies. Horseradish peroxidase-conjugated β-actin (SC-47778, Santa Cruz Biotechnology, USA) and antibodies against ATG7 (#8558, Cell Signaling Technology, USA), LC3 (NB100-2220, Novus Biologicals, USA), p62 (P0067, Sigma-Aldrich, USA), sex-determining region Y-box 2 (Sox2, ab97959, Abcam, UK), microtubule-associated protein 2 (MAP2, ab5392, Abcam), receptor interaction protein (RIP, MAB1580, Merck, USA), glial fibrillary acidic protein (GFAP, NBP1-05198, Novus Biologicals), nestin (bs-0008R-A555, Bioss, USA), and GFP (SC-9996, Santa Cruz Biotechnology) were purchased from the indicated companies.
HCN cells were maintained at 37℃, 5% CO2 on dishes coated with poly-L-ornithine (#3655, Sigma-Aldrich) and laminin (#35432, Corning, USA) in chemically composed HCN cell medium, as previously described [41,42]. For differentiation studies, HCN cells were plated onto coated dishes at a density of 1×105/cm2; 24 h later, medium was changed to differentiation medium and the cells were cultured for 4 days (Fig. 1A). Differentiation medium composition was Dulbecco's modified Eagle's medium/F-12 with the following additions: for neurons, 1 µM RA, 5 µM forskolin, 0.1% FBS; for oligodendrocytes, 1 µM RA, 2 ng/ml basic fibroblast growth factor, 1% FBS; for astrocytes, 1 µM RA, 5% FBS.
Lentiviral shRNA clones targeting rat Atg7 (TRCN0000092164, TRCN0000369085) were purchased from the Mission Library (Sigma-Aldrich). Lentiviruses were produced following published protocols and were used to infect HCN cells [43]. For stable expression of mRFP-GFP-LC3, HCN cells were infected with lentivirus expressing pLjm1-mRFP-GFP-LC3. For stable knockdown or overexpression, HCN cells were infected with the virus for 24 h and then the medium was replaced with fresh medium. After 72 h, HCN cells were treated with puromycin (5 µg/ml) for 6 h and then maintained in medium containing puromycin (1 µg/ml). The rat p62 (AGCTGAAGCGGCGGATCTCGCGG) single guide (sg) RNA was designed by using an online program (http://crispr.mit.edu) and cloned into the plenti-CRISPR-v2 vector (#52961, Addgene, USA).
Cells were rinsed with phosphate-buffered saline (PBS), and cell lysis and RNA isolation were performed using the QIAzol Lysis Reagent (#79306, Qiagen, Germany) following the manufacturer's instructions. cDNA was synthesized using the ImProm-II Reverse Transcriptase kit (#A3800, Promega, USA) and oligo dT primers, and was used for qPCR with TOPreal qPCR 2X PreMIX (#RT500, Enzynomics, Korea) and the following primers: rat
Cells were rinsed in PBS, fixed in 4% paraformaldehyde for 10 min, rinsed twice with PBS, and blocked for 5 min at room temperature in 0.2% Triton X-100 in antibody diluent solution (#003218, Thermo Fisher Scientific, USA); Triton X-100 was added for permeabilization. Cells were incubated with primary antibodies diluted in antibody diluent solution overnight at 4℃ and then with secondary antibodies prepared in antibody dilution solution for 1 h at room temperature. Secondary antibodies conjugated to Alexa Fluor 488 (#A11034, Thermo Fisher Scientific) or 647 (#703-605-155, Jackson Immuno Research Laboratories, USA) were used to visualize primary antibodies. Following incubation with antibodies, cells were rinsed twice with PBS, and the nuclei were stained for 10 min with Hoechst 33342 (#H3570, Thermo Fisher Scientific) at a 1:1000 dilution. Fluorescence images were obtained under an LSM700 or 780 confocal microscope (Carl Zeiss, Germany) and analyzed in Zen software (Carl Zeiss).
Cells were harvested and lysed for 30 min on ice in lysis buffer (50 mM Tris-HCl, pH 7.5, 250 mM sucrose, 1 mM EDTA, 1 mM EGTA, 1 mM dithiothreitol, 50 mM NaF, 1 mM phenylmethylsulfonyl fluoride, 1 mM benzamidine, 1% Triton X-100, supplemented with protease and phosphatase inhibitor cocktails (#78429, #1862495, Thermo Fisher Scientific). The samples were run on an SDS-polyacrylamide gel and transferred to a polyvinylidene fluoride membrane in a semi-dry electrophoretic transfer cell (Bio-Rad). Membranes were blocked with 5% nonfat dry milk powder dissolved in Tris-buffered saline with 0.1% Tween 20 (TBST) for 1 h at room temperature. The membranes were then incubated with appropriate primary antibodies overnight at 4℃ in a shaking incubator. Next day, membranes were washed with TBST 3 times, 10 min each and incubated with peroxidase-conjugated secondary antibodies diluted in blocking solution for 1 h at room temperature. After washing, proteins of interest were detected using a chemiluminescence detection kit (Thermo Fisher Scientific).
HCN cells were seeded in a 96-well plate with 1.0×105 cells per well and stained with propidium iodide (PI) (P4170, Sigma-Aldrich) and Hoechst 33342 (P3566, Invitrogen) at the indicated time points. The stock solutions of PI (10 mg/ml) and Hoechst (1 mg/ml) were diluted 1:1000 in PBS, and after incubation in the dark at 37℃ for 20 min, blue and red signal positive cells were counted under a fluorescence microscope (Axiovert 40 CFL, Carl Zeiss). Stained cells were quantified using automated image analysis software Matlab with CellC package. The percentage of cell death was calculated as follows:
Cell death (%)=[PI (red) positive cell number/ Hoechst (blue) positive cell number]×100.
All values are presented as mean±standard error of the mean (SEM) obtained by averaging the data from at least three independent experiments. Statistical significance was determined by a two-tailed unpaired Student's t-test using GraphPad Prism (GraphPad Software, USA).
RESULTS
To confirm their potential to differentiate into neurons, astrocytes, and oligodendrocytes, HCN cells were exposed to different differentiation media (Fig. 1A). Immunocytochemistry data showed that undifferentiated HCN cells highly expressed nestin, an NSC-specific marker (Fig 1B). After differentiation into neurons, oligodendrocytes, and astrocytes, the expression of respective lineage-specific markers (MAP2, mature neuronal marker; RIP, oligodendrocyte marker; GFAP, astrocyte marker) was readily detectable by immunofluorescent staining (Fig. 1B). We also performed qPCR to compare the mRNA levels of these markers between undifferentiated and differentiated HCN cells. First, we chose β-actin for normalization; however, its expression changed substantially during differentiation, possibly due to rapid remodeling of cytoskeleton (data not shown). Therefore, the gene for 18S ribosomal RNA (
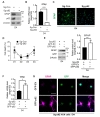
To study the role of autophagy during astrogenesis, we characterized the astrogenesis every 24 h (Fig. 2A). Before induction of differentiation, all HCN cells were nestin-positive, whereas GFAP was barely detectable. However, GFAP was detected in most cells while nestin was rarely detectable after the completion of astrogenesis (Fig. 2B). Time course analysis of GFAP immunoreactivity and protein levels showed a steady increase in the amount of GFAP during astrogenesis with a concurrent decrease in Sox2 (Fig. 2C and 2D). Cell morphology also changed from spherical stem cell shape to radial, which is typical for astrocytes (Fig. 2C). Consistent with the results of Western blotting analysis, mRNA levels of
Highly dynamic nature of neural development implies reconstitution of cellular components and remodeling of cellular structure during differentiation, requiring energy supply and participation of catabolic processes. To explore whether autophagy is activated in HCN cells during astrogenesis, we examined autophagosome formation in mRFP-GFP-LC3 stable cell lines. Before differentiation, the number of LC3 puncta was low in HCN cells. Interestingly, the number of LC3 puncta increased at early time points (D1 and D2), and decreased afterwards (D3 and D4) (Fig. 3A). Blocking autophagy flux with Baf.A1 should increase accumulation of LC3-II if autophagy flux rate is high [6]. Administration of Baf.A1 at D2 further increased autophagosome formation, suggesting that autophagic flux was very high at this time point (Fig. 3B). Western blotting analysis showed a similar increase in LC3-II and decrease in p62 at D1 and D2 of differentiation; their levels then returned to the basal state (Fig. 3C). To compare the autophagic flux between different time points, we treated the cells with Baf.A1 at D0, D2 and D4, and examined the LC3-II levels by Western blotting analysis. Interestingly, Baf.A1 treatment at D4 led to more significant accumulation of LC3-II than D2, although it did not reach statistical significance (Fig. 3D). These data suggest that although autophagy flux seems to return to the basal state after the peak at D2, the overall capacity of on-rate autophagy flux, which can be revealed after blocking of late stage of autophagy, still remains high until D4 during differentiation. In line with the assumption that autophagy was induced from early times points, Baf.A1 increased the levels of LC3-II at D2 (Fig. 3E). In addition to Baf.A1, we tested another autophagy blocker, pepstatin A and E64d (PepA/E64d), which inhibit lysosomal proteases [46]. Administration of PepA/E64d also increased LC3-II and p62 levels at D2 in a similar manner as Baf.A1 (Fig. 3E). These results suggest that autophagy flux was dramatically increased from D1 and D2 upon the induction of astrogenesis and seemingly declined afterwards at basal state. However, overall capacity of autophagy remained high until D4 stage.
To examine whether the upregulated autophagy is crucial for HCN cell differentiation into astrocytes, we genetically repressed autophagy by stable knockdown of Atg7 in HCN cells (Fig. 4A). As Atg7 plays a key role in autophagosome formation [47], its knockdown prevents autophagy in HCN cells, as we previously demonstrated [38,42,48]. Western blotting analysis showed a substantial reduction in the GFAP level in HCN cells with stable knockdown of Atg7 (Sh-Atg7) in comparison with control shRNA-transduced HCN cells (Sh-Con) (Fig. 4B). qPCR results also indicated a decrease in the
To confirm the requirement of autophagy for astrogenesis, we deleted another key autophagy-related gene,
DISCUSSION
Various intrinsic and extrinsic cues regulate astrocyte differentiation in the developing brain [50]. Being a major process for turnover and recycling of cellular constituents, autophagy is also intimately involved in development; thus, it is not surprising that autophagy is critical for the development of the nervous system. The role of autophagy in astrocyte differentiation in the developing brain was recently studied [51]. However, so far, the role of autophagy in astrocyte differentiation from adult NSCs is very little known. Although the present study was restricted to
Interestingly, we observed an increase in autophagy flux at relatively early time points during 5-day differentiation of HCN cells into astrocytes. Autophagy flux peaked on days 1 and 2, and then returned to a level similar to that in control HCN cells. However, when fusion of autophagosomes with lysosomes was blocked by Baf.A1, differentiating HCN cells at D4 still displayed similar or higher capacity of autophagy flux, compared with D2. When autophagy was suppressed by a stable knockdown of Atg7 or LC3 using sh-Atg7 or sh-LC3-expressing lentivirus or by genetic knockout of
In the presence of differentiation cues, autophagy stimulates differentiation of stem cells by regulating a dynamic and highly inducible metabolic process [55]. Autophagy is also a crucial player in intracellular remodeling, and this role of autophagy is particularly important for stem cells. Our results show that autophagy is critical for remodeling of adult NSCs. Due to high metabolic demand during differentiation and tissue remodeling, mitochondria are also important for differentiation and tissue remodeling. We previously reported that mitophagy occurs in HCN cells [38]. Autophagy may also regulate astrogenesis of HCN cells by controlling the mitochondrial network during differentiation [56].
Differentiation capability of neural stem/progenitor cells can be used for the development of treatments for various neurodegenerative diseases [57]. Studies using HCN cells can contribute to the molecular dissection of autophagy-mediated neural differentiation and better understanding of the basic and applied biology of adult NSCs.
Figures
References
- Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science 2000;290:1717-1721.
- Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, Griffiths G, Ktistakis NT. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol 2008;182:685-701.
- Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J 2000;19:5720-5728.
- Tanida I, Minematsu-Ikeguchi N, Ueno T, Kominami E. Lysosomal turnover, but not a cellular level, of endogenous LC3 is a marker for autophagy. Autophagy 2005;1:84-91.
- Mizushima N. Autophagy: process and function. Genes Dev 2007;21:2861-2873.
- Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Acevedo Arozena A, Adachi H, Adams CM, Adams PD, Adeli K, Adhihetty PJ, Adler SG, Agam G, Agarwal R, Aghi MK, Agnello M, Agostinis P, Aguilar PV, Aguirre-Ghiso J, Airoldi EM, Ait-Si-Ali S, Akematsu T, Akporiaye ET, Al-Rubeai M, Albaiceta GM, Albanese C, Albani D, Albert ML, Aldudo J, Algül H, Alirezaei M, Alloza I, Almasan A, Almonte-Beceril M, Alnemri ES, Alonso C, Altan-Bonnet N, Altieri DC, Alvarez S, Alvarez-Erviti L, Alves S, Amadoro G, Amano A, Amantini C, Ambrosio S, Amelio I, Amer AO, Amessou M, Amon A, An Z, Anania FA, Andersen SU, Andley UP, Andreadi CK, Andrieu-Abadie N, Anel A, Ann DK, Anoopkumar-Dukie S, Antonioli M, Aoki H, Apostolova N, Aquila S, Aquilano K, Araki K, Arama E, Aranda A, Araya J, Arcaro A, Arias E, Arimoto H, Ariosa AR, Armstrong JL, Arnould T, Arsov I, Asanuma K, Askanas V, Asselin E, Atarashi R, Atherton SS, Atkin JD, Attardi LD, Auberger P, Auburger G, Aurelian L, Autelli R, Avagliano L, Avantaggiati ML, Avrahami L, Awale S, Azad N, Bachetti T, Backer JM, Bae DH, Bae JS, Bae ON, Bae SH, Baehrecke EH, Baek SH, Baghdiguian S, Bagniewska-Zadworna A, Bai H, Bai J, Bai XY, Bailly Y, Balaji KN, Balduini W, Ballabio A, Balzan R, Banerjee R, Bánhegyi G, Bao H, Barbeau B, Barrachina MD, Barreiro E, Bartel B, Bartolomé A, Bassham DC, Bassi MT, Bast RC, Basu A, Batista MT, Batoko H, Battino M, Bauckman K, Baumgarner BL, Bayer KU, Beale R, Beaulieu JF, Beck GR, Becker C, Beckham JD, Bédard PA, Bednarski PJ, Begley TJ, Behl C, Behrends C, Behrens GM, Behrns KE, Bejarano E, Belaid A, Belleudi F, Bénard G, Berchem G, Bergamaschi D, Bergami M, Berkhout B, Berliocchi L, Bernard A, Bernard M, Bernassola F, Bertolotti A, Bess AS, Besteiro S, Bettuzzi S, Bhalla S, Bhattacharyya S, Bhutia SK, Biagosch C, Bianchi MW, Biard-Piechaczyk M, Billes V, Bincoletto C, Bingol B, Bird SW, Bitoun M, Bjedov I, Blackstone C, Blanc L, Blanco GA, Blomhoff HK, Boada-Romero E, Böckler S, Boes M, Boesze-Battaglia K, Boise LH, Bolino A, Boman A, Bonaldo P, Bordi M, Bosch J, Botana LM, Botti J, Bou G, Bouché M, Bouchecareilh M, Boucher MJ, Boulton ME, Bouret SG, Boya P, Boyer-Guittaut M, Bozhkov PV, Brady N, Braga VM, Brancolini C, Braus GH, Bravo-San Pedro JM, Brennan LA, Bresnick EH, Brest P, Bridges D, Bringer MA, Brini M, Brito GC, Brodin B, Brookes PS, Brown EJ, Brown K, Broxmeyer HE, Bruhat A, Brum PC, Brumell JH, Brunetti-Pierri N, Bryson-Richardson RJ, Buch S, Buchan AM, Budak H, Bulavin DV, Bultman SJ, Bultynck G, Bumbasirevic V, Burelle Y, Burke RE, Burmeister M, Bütikofer P, Caberlotto L, Cadwell K, Cahova M, Cai D, Cai J, Cai Q, Calatayud S, Camougrand N, Campanella M, Campbell GR, Campbell M, Campello S, Candau R, Caniggia I, Cantoni L, Cao L, Caplan AB, Caraglia M, Cardinali C, Cardoso SM, Carew JS, Carleton LA, Carlin CR, Carloni S, Carlsson SR, Carmona-Gutierrez D, Carneiro LA, Carnevali O, Carra S, Carrier A, Carroll B, Casas C, Casas J, Cassinelli G, Castets P, Castro-Obregon S, Cavallini G, Ceccherini I, Cecconi F, Cederbaum AI, Ceña V, Cenci S, Cerella C, Cervia D, Cetrullo S, Chaachouay H, Chae HJ, Chagin AS, Chai CY, Chakrabarti G, Chamilos G, Chan EY, Chan MT, Chandra D, Chandra P, Chang CP, Chang RC, Chang TY, Chatham JC, Chatterjee S, Chauhan S, Che Y, Cheetham ME, Cheluvappa R, Chen CJ, Chen G, Chen GC, Chen G, Chen H, Chen JW, Chen JK, Chen M, Chen M, Chen P, Chen Q, Chen Q, Chen SD, Chen S, Chen SS, Chen W, Chen WJ, Chen WQ, Chen W, Chen X, Chen YH, Chen YG, Chen Y, Chen Y, Chen Y, Chen YJ, Chen YQ, Chen Y, Chen Z, Chen Z, Cheng A, Cheng CH, Cheng H, Cheong H, Cherry S, Chesney J, Cheung CH, Chevet E, Chi HC, Chi SG, Chiacchiera F, Chiang HL, Chiarelli R, Chiariello M, Chieppa M, Chin LS, Chiong M, Chiu GN, Cho DH, Cho SG, Cho WC, Cho YY, Cho YS, Choi AM, Choi EJ, Choi EK, Choi J, Choi ME, Choi SI, Chou TF, Chouaib S, Choubey D, Choubey V, Chow KC, Chowdhury K, Chu CT, Chuang TH, Chun T, Chung H, Chung T, Chung YL, Chwae YJ, Cianfanelli V, Ciarcia R, Ciechomska IA, Ciriolo MR, Cirone M, Claerhout S, Clague MJ, Clària J, Clarke PG, Clarke R, Clementi E, Cleyrat C, Cnop M, Coccia EM, Cocco T, Codogno P, Coers J, Cohen EE, Colecchia D, Coletto L, Coll NS, Colucci-Guyon E, Comincini S, Condello M, Cook KL, Coombs GH, Cooper CD, Cooper JM, Coppens I, Corasaniti MT, Corazzari M, Corbalan R, Corcelle-Termeau E, Cordero MD, Corral-Ramos C, Corti O, Cossarizza A, Costelli P, Costes S, Cotman SL, Coto-Montes A, Cottet S, Couve E, Covey LR, Cowart LA, Cox JS, Coxon FP, Coyne CB, Cragg MS, Craven RJ, Crepaldi T, Crespo JL, Criollo A, Crippa V, Cruz MT, Cuervo AM, Cuezva JM, Cui T, Cutillas PR, Czaja MJ, Czyzyk-Krzeska MF, Dagda RK, Dahmen U, Dai C, Dai W, Dai Y, Dalby KN, Dalla Valle L, Dalmasso G, D'Amelio M, Damme M, Darfeuille-Michaud A, Dargemont C, Darley-Usmar VM, Dasarathy S, Dasgupta B, Dash S, Dass CR, Davey HM, Davids LM, Dávila D, Davis RJ, Dawson TM, Dawson VL, Daza P, de Belleroche J, de Figueiredo P, de Figueiredo RC, de la Fuente J, De Martino L, De Matteis A, De Meyer GR, De Milito A, De Santi M, de Souza W, De Tata V, De Zio D, Debnath J, Dechant R, Decuypere JP, Deegan S, Dehay B, Del Bello B, Del Re DP, Delage-Mourroux R, Delbridge LM, Deldicque L, Delorme-Axford E, Deng Y, Dengjel J, Denizot M, Dent P, Der CJ, Deretic V, Derrien B, Deutsch E, Devarenne TP, Devenish RJ, Di Bartolomeo S, Di Daniele N, Di Domenico F, Di Nardo A, Di Paola S, Di Pietro A, Di Renzo L, DiAntonio A, Díaz-Araya G, Díaz-Laviada I, Diaz-Meco MT, Diaz-Nido J, Dickey CA, Dickson RC, Diederich M, Digard P, Dikic I, Dinesh-Kumar SP, Ding C, Ding WX, Ding Z, Dini L, Distler JH, Diwan A, Djavaheri-Mergny M, Dmytruk K, Dobson RC, Doetsch V, Dokladny K, Dokudovskaya S, Donadelli M, Dong XC, Dong X, Dong Z, Donohue TM, Doran KS, D'Orazi G, Dorn GW, Dosenko V, Dridi S, Drucker L, Du J, Du LL, Du L, du Toit A, Dua P, Duan L, Duann P, Dubey VK, Duchen MR, Duchosal MA, Duez H, Dugail I, Dumit VI, Duncan MC, Dunlop EA, Dunn WA, Dupont N, Dupuis L, Durán RV, Durcan TM, Duvezin-Caubet S, Duvvuri U, Eapen V, Ebrahimi-Fakhari D, Echard A, Eckhart L, Edelstein CL, Edinger AL, Eichinger L, Eisenberg T, Eisenberg-Lerner A, Eissa NT, El-Deiry WS, El-Khoury V, Elazar Z, Eldar-Finkelman H, Elliott CJ, Emanuele E, Emmenegger U, Engedal N, Engelbrecht AM, Engelender S, Enserink JM, Erdmann R, Erenpreisa J, Eri R, Eriksen JL, Erman A, Escalante R, Eskelinen EL, Espert L, Esteban-Martínez L, Evans TJ, Fabri M, Fabrias G, Fabrizi C, Facchiano A, Færgeman NJ, Faggioni A, Fairlie WD, Fan C, Fan D, Fan J, Fang S, Fanto M, Fanzani A, Farkas T, Faure M, Favier FB, Fearnhead H, Federici M, Fei E, Felizardo TC, Feng H, Feng Y, Feng Y, Ferguson TA, Fernández ÁF, Fernandez-Barrena MG, Fernandez-Checa JC, Fernández-López A, Fernandez-Zapico ME, Feron O, Ferraro E, Ferreira-Halder CV, Fesus L, Feuer R, Fiesel FC, Filippi-Chiela EC, Filomeni G, Fimia GM, Fingert JH, Finkbeiner S, Finkel T, Fiorito F, Fisher PB, Flajolet M, Flamigni F, Florey O, Florio S, Floto RA, Folini M, Follo C, Fon EA, Fornai F, Fortunato F, Fraldi A, Franco R, Francois A, François A, Frankel LB, Fraser ID, Frey N, Freyssenet DG, Frezza C, Friedman SL, Frigo DE, Fu D, Fuentes JM, Fueyo J, Fujitani Y, Fujiwara Y, Fujiya M, Fukuda M, Fulda S, Fusco C, Gabryel B, Gaestel M, Gailly P, Gajewska M, Galadari S, Galili G, Galindo I, Galindo MF, Galliciotti G, Galluzzi L, Galluzzi L, Galy V, Gammoh N, Gandy S, Ganesan AK, Ganesan S, Ganley IG, Gannagé M, Gao FB, Gao F, Gao JX, García Nannig L, García Véscovi E, Garcia-Macía M, Garcia-Ruiz C, Garg AD, Garg PK, Gargini R, Gassen NC, Gatica D, Gatti E, Gavard J, Gavathiotis E, Ge L, Ge P, Ge S, Gean PW, Gelmetti V, Genazzani AA, Geng J, Genschik P, Gerner L, Gestwicki JE, Gewirtz DA, Ghavami S, Ghigo E, Ghosh D, Giammarioli AM, Giampieri F, Giampietri C, Giatromanolaki A, Gibbings DJ, Gibellini L, Gibson SB, Ginet V, Giordano A, Giorgini F, Giovannetti E, Girardin SE, Gispert S, Giuliano S, Gladson CL, Glavic A, Gleave M, Godefroy N, Gogal RM, Gokulan K, Goldman GH, Goletti D, Goligorsky MS, Gomes AV, Gomes LC, Gomez H, Gomez-Manzano C, Gómez-Sánchez R, Gonçalves DA, Goncu E, Gong Q, Gongora C, Gonzalez CB, Gonzalez-Alegre P, Gonzalez-Cabo P, González-Polo RA, Goping IS, Gorbea C, Gorbunov NV, Goring DR, Gorman AM, Gorski SM, Goruppi S, Goto-Yamada S, Gotor C, Gottlieb RA, Gozes I, Gozuacik D, Graba Y, Graef M, Granato GE, Grant GD, Grant S, Gravina GL, Green DR, Greenhough A, Greenwood MT, Grimaldi B, Gros F, Grose C, Groulx JF, Gruber F, Grumati P, Grune T, Guan JL, Guan KL, Guerra B, Guillen C, Gulshan K, Gunst J, Guo C, Guo L, Guo M, Guo W, Guo XG, Gust AA, Gustafsson ÅB, Gutierrez E, Gutierrez MG, Gwak HS, Haas A, Haber JE, Hadano S, Hagedorn M, Hahn DR, Halayko AJ, Hamacher-Brady A, Hamada K, Hamai A, Hamann A, Hamasaki M, Hamer I, Hamid Q, Hammond EM, Han F, Han W, Handa JT, Hanover JA, Hansen M, Harada M, Harhaji-Trajkovic L, Harper JW, Harrath AH, Harris AL, Harris J, Hasler U, Hasselblatt P, Hasui K, Hawley RG, Hawley TS, He C, He CY, He F, He G, He RR, He XH, He YW, He YY, Heath JK, Hébert MJ, Heinzen RA, Helgason GV, Hensel M, Henske EP, Her C, Herman PK, Hernández A, Hernandez C, Hernández-Tiedra S, Hetz C, Hiesinger PR, Higaki K, Hilfiker S, Hill BG, Hill JA, Hill WD, Hino K, Hofius D, Hofman P, Höglinger GU, Höhfeld J, Holz MK, Hong Y, Hood DA, Hoozemans JJ, Hoppe T, Hsu C, Hsu CY, Hsu LC, Hu D, Hu G, Hu HM, Hu H, Hu MC, Hu YC, Hu ZW, Hua F, Hua Y, Huang C, Huang HL, Huang KH, Huang KY, Huang S, Huang S, Huang WP, Huang YR, Huang Y, Huang Y, Huber TB, Huebbe P, Huh WK, Hulmi JJ, Hur GM, Hurley JH, Husak Z, Hussain SN, Hussain S, Hwang JJ, Hwang S, Hwang TI, Ichihara A, Imai Y, Imbriano C, Inomata M, Into T, Iovane V, Iovanna JL, Iozzo RV, Ip NY, Irazoqui JE, Iribarren P, Isaka Y, Isakovic AJ, Ischiropoulos H, Isenberg JS, Ishaq M, Ishida H, Ishii I, Ishmael JE, Isidoro C, Isobe K, Isono E, Issazadeh-Navikas S, Itahana K, Itakura E, Ivanov AI, Iyer AK, Izquierdo JM, Izumi Y, Izzo V, Jäättelä M, Jaber N, Jackson DJ, Jackson WT, Jacob TG, Jacques TS, Jagannath C, Jain A, Jana NR, Jang BK, Jani A, Janji B, Jannig PR, Jansson PJ, Jean S, Jendrach M, Jeon JH, Jessen N, Jeung EB, Jia K, Jia L, Jiang H, Jiang H, Jiang L, Jiang T, Jiang X, Jiang X, Jiang X, Jiang Y, Jiang Y, Jiménez A, Jin C, Jin H, Jin L, Jin M, Jin S, Jinwal UK, Jo EK, Johansen T, Johnson DE, Johnson GV, Johnson JD, Jonasch E, Jones C, Joosten LA, Jordan J, Joseph AM, Joseph B, Joubert AM, Ju D, Ju J, Juan HF, Juenemann K, Juhász G, Jung HS, Jung JU, Jung YK, Jungbluth H, Justice MJ, Jutten B, Kaakoush NO, Kaarniranta K, Kaasik A, Kabuta T, Kaeffer B, Kågedal K, Kahana A, Kajimura S, Kakhlon O, Kalia M, Kalvakolanu DV, Kamada Y, Kambas K, Kaminskyy VO, Kampinga HH, Kandouz M, Kang C, Kang R, Kang TC, Kanki T, Kanneganti TD, Kanno H, Kanthasamy AG, Kantorow M, Kaparakis-Liaskos M, Kapuy O, Karantza V, Karim MR, Karmakar P, Kaser A, Kaushik S, Kawula T, Kaynar AM, Ke PY, Ke ZJ, Kehrl JH, Keller KE, Kemper JK, Kenworthy AK, Kepp O, Kern A, Kesari S, Kessel D, Ketteler R, Kettelhut IC, Khambu B, Khan MM, Khandelwal VK, Khare S, Kiang JG, Kiger AA, Kihara A, Kim AL, Kim CH, Kim DR, Kim DH, Kim EK, Kim HY, Kim HR, Kim JS, Kim JH, Kim JC, Kim JH, Kim KW, Kim MD, Kim MM, Kim PK, Kim SW, Kim SY, Kim YS, Kim Y, Kimchi A, Kimmelman AC, Kimura T, King JS, Kirkegaard K, Kirkin V, Kirshenbaum LA, Kishi S, Kitajima Y, Kitamoto K, Kitaoka Y, Kitazato K, Kley RA, Klimecki WT, Klinkenberg M, Klucken J, Knævelsrud H, Knecht E, Knuppertz L, Ko JL, Kobayashi S, Koch JC, Koechlin-Ramonatxo C, Koenig U, Koh YH, Köhler K, Kohlwein SD, Koike M, Komatsu M, Kominami E, Kong D, Kong HJ, Konstantakou EG, Kopp BT, Korcsmaros T, Korhonen L, Korolchuk VI, Koshkina NV, Kou Y, Koukourakis MI, Koumenis C, Kovács AL, Kovács T, Kovacs WJ, Koya D, Kraft C, Krainc D, Kramer H, Kravic-Stevovic T, Krek W, Kretz-Remy C, Krick R, Krishnamurthy M, Kriston-Vizi J, Kroemer G, Kruer MC, Kruger R, Ktistakis NT, Kuchitsu K, Kuhn C, Kumar AP, Kumar A, Kumar A, Kumar D, Kumar D, Kumar R, Kumar S, Kundu M, Kung HJ, Kuno A, Kuo SH, Kuret J, Kurz T, Kwok T, Kwon TK, Kwon YT, Kyrmizi I, La Spada AR, Lafont F, Lahm T, Lakkaraju A, Lam T, Lamark T, Lancel S, Landowski TH, Lane DJ, Lane JD, Lanzi C, Lapaquette P, Lapierre LR, Laporte J, Laukkarinen J, Laurie GW, Lavandero S, Lavie L, LaVoie MJ, Law BY, Law HK, Law KB, Layfield R, Lazo PA, Le Cam L, Le Roch KG, Le Stunff H, Leardkamolkarn V, Lecuit M, Lee BH, Lee CH, Lee EF, Lee GM, Lee HJ, Lee H, Lee JK, Lee J, Lee JH, Lee JH, Lee M, Lee MS, Lee PJ, Lee SW, Lee SJ, Lee SJ, Lee SY, Lee SH, Lee SS, Lee SJ, Lee S, Lee YR, Lee YJ, Lee YH, Leeuwenburgh C, Lefort S, Legouis R, Lei J, Lei QY, Leib DA, Leibowitz G, Lekli I, Lemaire SD, Lemasters JJ, Lemberg MK, Lemoine A, Leng S, Lenz G, Lenzi P, Lerman LO, Lettieri Barbato D, Leu JI, Leung HY, Levine B, Lewis PA, Lezoualc'h F, Li C, Li F, Li FJ, Li J, Li K, Li L, Li M, Li M, Li Q, Li R, Li S, Li W, Li W, Li X, Li Y, Lian J, Liang C, Liang Q, Liao Y, Liberal J, Liberski PP, Lie P, Lieberman AP, Lim HJ, Lim KL, Lim K, Lima RT, Lin CS, Lin CF, Lin F, Lin F, Lin FC, Lin K, Lin KH, Lin PH, Lin T, Lin WW, Lin YS, Lin Y, Linden R, Lindholm D, Lindqvist LM, Lingor P, Linkermann A, Liotta LA, Lipinski MM, Lira VA, Lisanti MP, Liton PB, Liu B, Liu C, Liu CF, Liu F, Liu HJ, Liu J, Liu JJ, Liu JL, Liu K, Liu L, Liu L, Liu Q, Liu RY, Liu S, Liu S, Liu W, Liu XD, Liu X, Liu XH, Liu X, Liu X, Liu X, Liu Y, Liu Y, Liu Z, Liu Z, Liuzzi JP, Lizard G, Ljujic M, Lodhi IJ, Logue SE, Lokeshwar BL, Long YC, Lonial S, Loos B, López-Otín C, López-Vicario C, Lorente M, Lorenzi PL, Lõrincz P, Los M, Lotze MT, Lovat PE, Lu B, Lu B, Lu J, Lu Q, Lu SM, Lu S, Lu Y, Luciano F, Luckhart S, Lucocq JM, Ludovico P, Lugea A, Lukacs NW, Lum JJ, Lund AH, Luo H, Luo J, Luo S, Luparello C, Lyons T, Ma J, Ma Y, Ma Y, Ma Z, Machado J, Machado-Santelli GM, Macian F, MacIntosh GC, MacKeigan JP, Macleod KF, MacMicking JD, MacMillan-Crow LA, Madeo F, Madesh M, Madrigal-Matute J, Maeda A, Maeda T, Maegawa G, Maellaro E, Maes H, Magariños M, Maiese K, Maiti TK, Maiuri L, Maiuri MC, Maki CG, Malli R, Malorni W, Maloyan A, Mami-Chouaib F, Man N, Mancias JD, Mandelkow EM, Mandell MA, Manfredi AA, Manié SN, Manzoni C, Mao K, Mao Z, Mao ZW, Marambaud P, Marconi AM, Marelja Z, Marfe G, Margeta M, Margittai E, Mari M, Mariani FV, Marin C, Marinelli S, Mariño G, Markovic I, Marquez R, Martelli AM, Martens S, Martin KR, Martin SJ, Martin S, Martin-Acebes MA, Martín-Sanz P, Martinand-Mari C, Martinet W, Martinez J, Martinez-Lopez N, Martinez-Outschoorn U, Martínez-Velázquez M, Martinez-Vicente M, Martins WK, Mashima H, Mastrianni JA, Matarese G, Matarrese P, Mateo R, Matoba S, Matsumoto N, Matsushita T, Matsuura A, Matsuzawa T, Mattson MP, Matus S, Maugeri N, Mauvezin C, Mayer A, Maysinger D, Mazzolini GD, McBrayer MK, McCall K, McCormick C, McInerney GM, McIver SC, McKenna S, McMahon JJ, Mc-Neish IA, Mechta-Grigoriou F, Medema JP, Medina DL, Megyeri K, Mehrpour M, Mehta JL, Mei Y, Meier UC, Meijer AJ, Meléndez A, Melino G, Melino S, de Melo EJ, Mena MA, Meneghini MD, Menendez JA, Menezes R, Meng L, Meng LH, Meng S, Menghini R, Menko AS, Menna-Barreto RF, Menon MB, Meraz-Ríos MA, Merla G, Merlini L, Merlot AM, Meryk A, Meschini S, Meyer JN, Mi MT, Miao CY, Micale L, Michaeli S, Michiels C, Migliaccio AR, Mihailidou AS, Mijaljica D, Mikoshiba K, Milan E, Miller-Fleming L, Mills GB, Mills IG, Minakaki G, Minassian BA, Ming XF, Minibayeva F, Minina EA, Mintern JD, Minucci S, Miranda-Vizuete A, Mitchell CH, Miyamoto S, Miyazawa K, Mizushima N, Mnich K, Mograbi B, Mohseni S, Moita LF, Molinari M, Molinari M, Møller AB, Mollereau B, Mollinedo F, Mongillo M, Monick MM, Montagnaro S, Montell C, Moore DJ, Moore MN, Mora-Rodriguez R, Moreira PI, Morel E, Morelli MB, Moreno S, Morgan MJ, Moris A, Moriyasu Y, Morrison JL, Morrison LA, Morselli E, Moscat J, Moseley PL, Mostowy S, Motori E, Mottet D, Mottram JC, Moussa CE, Mpakou VE, Mukhtar H, Mulcahy Levy JM, Muller S, Muñoz-Moreno R, Muñoz-Pinedo C, Münz C, Murphy ME, Murray JT, Murthy A, Mysorekar IU, Nabi IR, Nabissi M, Nader GA, Nagahara Y, Nagai Y, Nagata K, Nagelkerke A, Nagy P, Naidu SR, Nair S, Nakano H, Nakatogawa H, Nanjundan M, Napolitano G, Naqvi NI, Nardacci R, Narendra DP, Narita M, Nascimbeni AC, Natarajan R, Navegantes LC, Nawrocki ST, Nazarko TY, Nazarko VY, Neill T, Neri LM, Netea MG, Netea-Maier RT, Neves BM, Ney PA, Nezis IP, Nguyen HT, Nguyen HP, Nicot AS, Nilsen H, Nilsson P, Nishimura M, Nishino I, Niso-Santano M, Niu H, Nixon RA, Njar VC, Noda T, Noegel AA, Nolte EM, Norberg E, Norga KK, Noureini SK, Notomi S, Notterpek L, Nowikovsky K, Nukina N, Nürnberger T, O'Donnell VB, O'Donovan T, O'Dwyer PJ, Oehme I, Oeste CL, Ogawa M, Ogretmen B, Ogura Y, Oh YJ, Ohmuraya M, Ohshima T, Ojha R, Okamoto K, Okazaki T, Oliver FJ, Ollinger K, Olsson S, Orban DP, Ordonez P, Orhon I, Orosz L, O'Rourke EJ, Orozco H, Ortega AL, Ortona E, Osellame LD, Oshima J, Oshima S, Osiewacz HD, Otomo T, Otsu K, Ou JH, Outeiro TF, Ouyang DY, Ouyang H, Overholtzer M, Ozbun MA, Ozdinler PH, Ozpolat B, Pacelli C, Paganetti P, Page G, Pages G, Pagnini U, Pajak B, Pak SC, Pakos-Zebrucka K, Pakpour N, Palková Z, Palladino F, Pallauf K, Pallet N, Palmieri M, Paludan SR, Palumbo C, Palumbo S, Pampliega O, Pan H, Pan W, Panaretakis T, Pandey A, Pantazopoulou A, Papackova Z, Papademetrio DL, Papassideri I, Papini A, Parajuli N, Pardo J, Parekh VV, Parenti G, Park JI, Park J, Park OK, Parker R, Parlato R, Parys JB, Parzych KR, Pasquet JM, Pasquier B, Pasumarthi KB, Patschan D, Patterson C, Pattingre S, Pattison S, Pause A, Pavenstädt H, Pavone F, Pedrozo Z, Peña FJ, Peñalva MA, Pende M, Peng J, Penna F, Penninger JM, Pensalfini A, Pepe S, Pereira GJ, Pereira PC, Pérez-de la Cruz V, Pérez-Pérez ME, Pérez-Rodríguez D, Pérez-Sala D, Perier C, Perl A, Perlmutter DH, Perrotta I, Pervaiz S, Pesonen M, Pessin JE, Peters GJ, Petersen M, Petrache I, Petrof BJ, Petrovski G, Phang JM, Piacentini M, Pierdominici M, Pierre P, Pierrefite-Carle V, Pietrocola F, Pimentel-Muiños FX, Pinar M, Pineda B, Pinkas-Kramarski R, Pinti M, Pinton P, Piperdi B, Piret JM, Platanias LC, Platta HW, Plowey ED, Pöggeler S, Poirot M, Polčic P, Poletti A, Poon AH, Popelka H, Popova B, Poprawa I, Poulose SM, Poulton J, Powers SK, Powers T, Pozuelo-Rubio M, Prak K, Prange R, Prescott M, Priault M, Prince S, Proia RL, Proikas-Cezanne T, Prokisch H, Promponas VJ, Przyklenk K, Puertollano R, Pugazhenthi S, Puglielli L, Pujol A, Puyal J, Pyeon D, Qi X, Qian WB, Qin ZH, Qiu Y, Qu Z, Quadrilatero J, Quinn F, Raben N, Rabinowich H, Radogna F, Ragusa MJ, Rahmani M, Raina K, Ramanadham S, Ramesh R, Rami A, Randall-Demllo S, Randow F, Rao H, Rao VA, Rasmussen BB, Rasse TM, Ratovitski EA, Rautou PE, Ray SK, Razani B, Reed BH, Reggiori F, Rehm M, Reichert AS, Rein T, Reiner DJ, Reits E, Ren J, Ren X, Renna M, Reusch JE, Revuelta JL, Reyes L, Rezaie AR, Richards RI, Richardson DR, Richetta C, Riehle MA, Rihn BH, Rikihisa Y, Riley BE, Rimbach G, Rippo MR, Ritis K, Rizzi F, Rizzo E, Roach PJ, Robbins J, Roberge M, Roca G, Roccheri MC, Rocha S, Rodrigues CM, Rodríguez CI, de Cordoba SR, Rodriguez-Muela N, Roelofs J, Rogov VV, Rohn TT, Rohrer B, Romanelli D, Romani L, Romano PS, Roncero MI, Rosa JL, Rosello A, Rosen KV, Rosenstiel P, Rost-Roszkowska M, Roth KA, Roué G, Rouis M, Rouschop KM, Ruan DT, Ruano D, Rubinsztein DC, Rucker EB, Rudich A, Rudolf E, Rudolf R, Ruegg MA, Ruiz-Roldan C, Ruparelia AA, Rusmini P, Russ DW, Russo GL, Russo G, Russo R, Rusten TE, Ryabovol V, Ryan KM, Ryter SW, Sabatini DM, Sacher M, Sachse C, Sack MN, Sadoshima J, Saftig P, Sagi-Eisenberg R, Sahni S, Saikumar P, Saito T, Saitoh T, Sakakura K, Sakoh-Nakatogawa M, Sakuraba Y, Salazar-Roa M, Salomoni P, Saluja AK, Salvaterra PM, Salvioli R, Samali A, Sanchez AM, Sánchez-Alcázar JA, Sanchez-Prieto R, Sandri M, Sanjuan MA, Santaguida S, Santambrogio L, Santoni G, Dos Santos CN, Saran S, Sardiello M, Sargent G, Sarkar P, Sarkar S, Sarrias MR, Sarwal MM, Sasakawa C, Sasaki M, Sass M, Sato K, Sato M, Satriano J, Savaraj N, Saveljeva S, Schaefer L, Schaible UE, Scharl M, Schatzl HM, Schekman R, Scheper W, Schiavi A, Schipper HM, Schmeisser H, Schmidt J, Schmitz I, Schneider BE, Schneider EM, Schneider JL, Schon EA, Schönenberger MJ, Schönthal AH, Schorderet DF, Schröder B, Schuck S, Schulze RJ, Schwarten M, Schwarz TL, Sciarretta S, Scotto K, Scovassi AI, Screaton RA, Screen M, Seca H, Sedej S, Segatori L, Segev N, Seglen PO, Seguí-Simarro JM, Segura-Aguilar J, Seki E, Sell C, Seiliez I, Semenkovich CF, Semenza GL, Sen U, Serra AL, Serrano-Puebla A, Sesaki H, Setoguchi T, Settembre C, Shacka JJ, Shajahan-Haq AN, Shapiro IM, Sharma S, She H, Shen CK, Shen CC, Shen HM, Shen S, Shen W, Sheng R, Sheng X, Sheng ZH, Shepherd TG, Shi J, Shi Q, Shi Q, Shi Y, Shibutani S, Shibuya K, Shidoji Y, Shieh JJ, Shih CM, Shimada Y, Shimizu S, Shin DW, Shinohara ML, Shintani M, Shintani T, Shioi T, Shirabe K, Shiri-Sverdlov R, Shirihai O, Shore GC, Shu CW, Shukla D, Sibirny AA, Sica V, Sigurdson CJ, Sigurdsson EM, Sijwali PS, Sikorska B, Silveira WA, Silvente-Poirot S, Silverman GA, Simak J, Simmet T, Simon AK, Simon HU, Simone C, Simons M, Simonsen A, Singh R, Singh SV, Singh SK, Sinha D, Sinha S, Sinicrope FA, Sirko A, Sirohi K, Sishi BJ, Sittler A, Siu PM, Sivridis E, Skwarska A, Slack R, Slaninová I, Slavov N, Smaili SS, Smalley KS, Smith DR, Soenen SJ, Soleimanpour SA, Solhaug A, Somasundaram K, Son JH, Sonawane A, Song C, Song F, Song HK, Song JX, Song W, Soo KY, Sood AK, Soong TW, Soontornniyomkij V, Sorice M, Sotgia F, Soto-Pantoja DR, Sotthibundhu A, Sousa MJ, Spaink HP, Span PN, Spang A, Sparks JD, Speck PG, Spector SA, Spies CD, Springer W, Clair DS, Stacchiotti A, Staels B, Stang MT, Starczynowski DT, Starokadomskyy P, Steegborn C, Steele JW, Stefanis L, Steffan J, Stellrecht CM, Stenmark H, Stepkowski TM, Stern ST, Stevens C, Stockwell BR, Stoka V, Storchova Z, Stork B, Stratoulias V, Stravopodis DJ, Strnad P, Strohecker AM, Ström AL, Stromhaug P, Stulik J, Su YX, Su Z, Subauste CS, Subramaniam S, Sue CM, Suh SW, Sui X, Sukseree S, Sulzer D, Sun FL, Sun J, Sun J, Sun SY, Sun Y, Sun Y, Sun Y, Sundaramoorthy V, Sung J, Suzuki H, Suzuki K, Suzuki N, Suzuki T, Suzuki YJ, Swanson MS, Swanton C, Swärd K, Swarup G, Sweeney ST, Sylvester PW, Szatmari Z, Szegezdi E, Szlosarek PW, Taegtmeyer H, Tafani M, Taillebourg E, Tait SW, Takacs-Vellai K, Takahashi Y, Takáts S, Takemura G, Takigawa N, Talbot NJ, Tamagno E, Tamburini J, Tan CP, Tan L, Tan ML, Tan M, Tan YJ, Tanaka K, Tanaka M, Tang D, Tang D, Tang G, Tanida I, Tanji K, Tannous BA, Tapia JA, Tasset-Cuevas I, Tatar M, Tavassoly I, Tavernarakis N, Taylor A, Taylor GS, Taylor GA, Taylor JP, Taylor MJ, Tchetina EV, Tee AR, Teixeira-Clerc F, Telang S, Tencomnao T, Teng BB, Teng RJ, Terro F, Tettamanti G, Theiss AL, Theron AE, Thomas KJ, Thomé MP, Thomes PG, Thorburn A, Thorner J, Thum T, Thumm M, Thurston TL, Tian L, Till A, Ting JP, Titorenko VI, Toker L, Toldo S, Tooze SA, Topisirovic I, Torgersen ML, Torosantucci L, Torriglia A, Torrisi MR, Tournier C, Towns R, Trajkovic V, Travassos LH, Triola G, Tripathi DN, Trisciuoglio D, Troncoso R, Trougakos IP, Truttmann AC, Tsai KJ, Tschan MP, Tseng YH, Tsukuba T, Tsung A, Tsvetkov AS, Tu S, Tuan HY, Tucci M, Tumbarello DA, Turk B, Turk V, Turner RF, Tveita AA, Tyagi SC, Ubukata M, Uchiyama Y, Udelnow A, Ueno T, Umekawa M, Umemiya-Shirafuji R, Underwood BR, Ungermann C, Ureshino RP, Ushioda R, Uversky VN, Uzcátegui NL, Vaccari T, Vaccaro MI, Váchová L, Vakifahmetoglu-Norberg H, Valdor R, Valente EM, Vallette F, Valverde AM, Van den Berghe G, Van Den Bosch L, van den Brink GR, van der Goot FG, van der Klei IJ, van der Laan LJ, van Doorn WG, van Egmond M, van Golen KL, Van Kaer L, van Lookeren Campagne M, Vandenabeele P, Vandenberghe W, Vanhorebeek I, Varela-Nieto I, Vasconcelos MH, Vasko R, Vavvas DG, Vega-Naredo I, Velasco G, Velentzas AD, Velentzas PD, Vellai T, Vellenga E, Vendelbo MH, Venkatachalam K, Ventura N, Ventura S, Veras PS, Verdier M, Vertessy BG, Viale A, Vidal M, Vieira HL, Vierstra RD, Vigneswaran N, Vij N, Vila M, Villar M, Villar VH, Villarroya J, Vindis C, Viola G, Viscomi MT, Vitale G, Vogl DT, Voitsekhovskaja OV, von Haefen C, von Schwarzenberg K, Voth DE, Vouret-Craviari V, Vuori K, Vyas JM, Waeber C, Walker CL, Walker MJ, Walter J, Wan L, Wan X, Wang B, Wang C, Wang CY, Wang C, Wang C, Wang C, Wang D, Wang F, Wang F, Wang G, Wang HJ, Wang H, Wang HG, Wang H, Wang HD, Wang J, Wang J, Wang M, Wang MQ, Wang PY, Wang P, Wang RC, Wang S, Wang TF, Wang X, Wang XJ, Wang XW, Wang X, Wang X, Wang Y, Wang Y, Wang Y, Wang YJ, Wang Y, Wang Y, Wang YT, Wang Y, Wang ZN, Wappner P, Ward C, Ward DM, Warnes G, Watada H, Watanabe Y, Watase K, Weaver TE, Weekes CD, Wei J, Weide T, Weihl CC, Weindl G, Weis SN, Wen L, Wen X, Wen Y, Westermann B, Weyand CM, White AR, White E, Whitton JL, Whitworth AJ, Wiels J, Wild F, Wildenberg ME, Wileman T, Wilkinson DS, Wilkinson S, Willbold D, Williams C, Williams K, Williamson PR, Winklhofer KF, Witkin SS, Wohlgemuth SE, Wollert T, Wolvetang EJ, Wong E, Wong GW, Wong RW, Wong VK, Woodcock EA, Wright KL, Wu C, Wu D, Wu GS, Wu J, Wu J, Wu M, Wu M, Wu S, Wu WK, Wu Y, Wu Z, Xavier CP, Xavier RJ, Xia GX, Xia T, Xia W, Xia Y, Xiao H, Xiao J, Xiao S, Xiao W, Xie CM, Xie Z, Xie Z, Xilouri M, Xiong Y, Xu C, Xu C, Xu F, Xu H, Xu H, Xu J, Xu J, Xu J, Xu L, Xu X, Xu Y, Xu Y, Xu ZX, Xu Z, Xue Y, Yamada T, Yamamoto A, Yamanaka K, Yamashina S, Yamashiro S, Yan B, Yan B, Yan X, Yan Z, Yanagi Y, Yang DS, Yang JM, Yang L, Yang M, Yang PM, Yang P, Yang Q, Yang W, Yang WY, Yang X, Yang Y, Yang Y, Yang Z, Yang Z, Yao MC, Yao PJ, Yao X, Yao Z, Yao Z, Yasui LS, Ye M, Yedvobnick B, Yeganeh B, Yeh ES, Yeyati PL, Yi F, Yi L, Yin XM, Yip CK, Yoo YM, Yoo YH, Yoon SY, Yoshida K, Yoshimori T, Young KH, Yu H, Yu JJ, Yu JT, Yu J, Yu L, Yu WH, Yu XF, Yu Z, Yuan J, Yuan ZM, Yue BY, Yue J, Yue Z, Zacks DN, Zacksenhaus E, Zaffaroni N, Zaglia T, Zakeri Z, Zecchini V, Zeng J, Zeng M, Zeng Q, Zervos AS, Zhang DD, Zhang F, Zhang G, Zhang GC, Zhang H, Zhang H, Zhang H, Zhang H, Zhang J, Zhang J, Zhang J, Zhang J, Zhang JP, Zhang L, Zhang L, Zhang L, Zhang L, Zhang MY, Zhang X, Zhang XD, Zhang Y, Zhang Y, Zhang Y, Zhang Y, Zhang Y, Zhao M, Zhao WL, Zhao X, Zhao YG, Zhao Y, Zhao Y, Zhao YX, Zhao Z, Zhao ZJ, Zheng D, Zheng XL, Zheng X, Zhivotovsky B, Zhong Q, Zhou GZ, Zhou G, Zhou H, Zhou SF, Zhou XJ, Zhu H, Zhu H, Zhu WG, Zhu W, Zhu XF, Zhu Y, Zhuang SM, Zhuang X, Ziparo E, Zois CE, Zoladek T, Zong WX, Zorzano A, Zughaier SM. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 2016;12:1-222.
- Kabeya Y, Mizushima N, Yamamoto A, Oshitani-Okamoto S, Ohsumi Y, Yoshimori T. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J Cell Sci 2004;117:2805-2812.
- Taherbhoy AM, Tait SW, Kaiser SE, Williams AH, Deng A, Nourse A, Hammel M, Kurinov I, Rock CO, Green DR, Schulman BA. Atg8 transfer from Atg7 to Atg3: a distinctive E1–E2 architecture and mechanism in the autophagy pathway. Mol Cell 2011;44:451-461.
- Mizushima N, Yoshimori T. How to interpret LC3 immunoblotting. Autophagy 2007;3:542-545.
- Tanida I, Ueno T, Kominami E. LC3 conjugation system in mammalian autophagy. Int J Biochem Cell Biol 2004;36:2503-2518.
- Bjørkøy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H, Johansen T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol 2005;171:603-614.
- Komatsu M, Waguri S, Koike M, Sou YS, Ueno T, Hara T, Mizushima N, Iwata J, Ezaki J, Murata S, Hamazaki J, Nishito Y, Iemura S, Natsume T, Yanagawa T, Uwayama J, Warabi E, Yoshida H, Ishii T, Kobayashi A, Yamamoto M, Yue Z, Uchiyama Y, Kominami E, Tanaka K. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell 2007;131:1149-1163.
- Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Øvervatn A, Bjørkøy G, Johansen T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem 2007;282:24131-24145.
- Berg TO, Fengsrud M, Strømhaug PE, Berg T, Seglen PO. Isolation and characterization of rat liver amphisomes. Evidence for fusion of autophagosomes with both early and late endosomes. J Biol Chem 1998;273:21883-21892.
- Yamamoto A, Tagawa Y, Yoshimori T, Moriyama Y, Masaki R, Tashiro Y. Bafilomycin A1 prevents maturation of autophagic vacuoles by inhibiting fusion between autophagosomes and lysosomes in rat hepatoma cell line, H-4-II-E cells. Cell Struct Funct 1998;23:33-42.
- Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy 2007;3:452-460.
- Mizushima N, Levine B. Autophagy in mammalian development and differentiation. Nat Cell Biol 2010;12:823-830.
- Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N. The role of autophagy during the early neonatal starvation period. Nature 2004;432:1032-1036.
- Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci U S A 2003;100:15077-15082.
- Tsukamoto S, Kuma A, Murakami M, Kishi C, Yamamoto A, Mizushima N. Autophagy is essential for preimplantation development of mouse embryos. Science 2008;321:117-120.
- Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E, Tanaka K. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 2006;441:880-884.
- Fimia GM, Stoykova A, Romagnoli A, Giunta L, Di Bartolomeo S, Nardacci R, Corazzari M, Fuoco C, Ucar A, Schwartz P, Gruss P, Piacentini M, Chowdhury K, Cecconi F. Ambra1 regulates autophagy and development of the nervous system. Nature 2007;447:1121-1125.
- Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, Mizushima N. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 2006;441:885-889.
- Vázquez P, Arroba AI, Cecconi F, de la Rosa EJ, Boya P, de Pablo F. Atg5 and Ambra1 differentially modulate neurogenesis in neural stem cells. Autophagy 2012;8:187-199.
- Wang C, Liang CC, Bian ZC, Zhu Y, Guan JL. FIP200 is required for maintenance and differentiation of postnatal neural stem cells. Nat Neurosci 2013;16:532-542.
- Fields RD, Araque A, Johansen-Berg H, Lim SS, Lynch G, Nave KA, Nedergaard M, Perez R, Sejnowski T, Wake H. Glial biology in learning and cognition. Neuroscientist 2014;20:426-431.
- Han X, Chen M, Wang F, Windrem M, Wang S, Shanz S, Xu Q, Oberheim NA, Bekar L, Betstadt S, Silva AJ, Takano T, Goldman SA, Nedergaard M. Forebrain engraftment by human glial progenitor cells enhances synaptic plasticity and learning in adult mice. Cell Stem Cell 2013;12:342-353.
- Clarke LE, Barres BA. Emerging roles of astrocytes in neural circuit development. Nat Rev Neurosci 2013;14:311-321.
- Jo S, Yarishkin O, Hwang YJ, Chun YE, Park M, Woo DH, Bae JY, Kim T, Lee J, Chun H, Park HJ, Lee DY, Hong J, Kim HY, Oh SJ, Park SJ, Lee H, Yoon BE, Kim Y, Jeong Y, Shim I, Bae YC, Cho J, Kowall NW, Ryu H, Hwang E, Kim D, Lee CJ. GABA from reactive astrocytes impairs memory in mouse models of Alzheimer's disease. Nat Med 2014;20:886-896.
- Lledo PM, Alonso M, Grubb MS. Adult neurogenesis and functional plasticity in neuronal circuits. Nat Rev Neurosci 2006;7:179-193.
- Gage FH. Neurogenesis in the adult brain. J Neurosci 2002;22:612-613.
- van Praag H, Schinder AF, Christie BR, Toni N, Palmer TD, Gage FH. Functional neurogenesis in the adult hippocampus. Nature 2002;415:1030-1034.
- Zhao C, Deng W, Gage FH. Mechanisms and functional implications of adult neurogenesis. Cell 2008;132:645-660.
- Suh H, Deng W, Gage FH. Signaling in adult neurogenesis. Annu Rev Cell Dev Biol 2009;25:253-275.
- Gage FH, Temple S. Neural stem cells: generating and regenerating the brain. Neuron 2013;80:588-601.
- Ming GL, Song H. Adult neurogenesis in the mammalian central nervous system. Annu Rev Neurosci 2005;28:223-250.
- Palmer TD, Takahashi J, Gage FH. The adult rat hippocampus contains primordial neural stem cells. Mol Cell Neurosci 1997;8:389-404.
- Ha S, Jeong SH, Yi K, Chung KM, Hong CJ, Kim SW, Kim EK, Yu SW. Phosphorylation of p62 by AMP-activated protein kinase mediates autophagic cell death in adult hippocampal neural stem cells. J Biol Chem 2017;292:13795-13808.
- Chung KM, Jeong EJ, Park H, An HK, Yu SW. Mediation of autophagic cell death by type 3 ryanodine receptor (RyR3) in adult hippocampal neural stem cells. Front Cell Neurosci 2016;10:116-130.
- Ha S, Ryu HY, Chung KM, Baek SH, Kim EK, Yu SW. Regulation of autophagic cell death by glycogen synthase kinase-3β in adult hippocampal neural stem cells following insulin withdrawal. Mol Brain 2015;8:30-41.
- Chung KM, Park H, Jung S, Ha S, Yoo SJ, Woo H, Lee HJ, Kim SW, Kim EK, Moon C, Yu SW. Calpain determines the propensity of adult hippocampal neural stem cells to autophagic cell death following insulin withdrawal. Stem Cells 2015;33:3052-3064.
- Yu SW, Baek SH, Brennan RT, Bradley CJ, Park SK, Lee YS, Jun EJ, Lookingland KJ, Kim EK, Lee H, Goudreau JL, Kim SW. Autophagic death of adult hippocampal neural stem cells following insulin withdrawal. Stem Cells 2008;26:2602-2610.
- Tiscornia G, Singer O, Verma IM. Production and purification of lentiviral vectors. Nat Protoc 2006;1:241-245.
- Götz M, Bolz J, Joester A, Faissner A. Tenascin-C synthesis and influence on axonal growth during rat cortical development. Eur J Neurosci 1997;9:496-506.
- Bartsch S, Bartsch U, Dörries U, Faissner A, Weller A, Ekblom P, Schachner M. Expression of tenascin in the developing and adult cerebellar cortex. J Neurosci 1992;12:736-749.
- Baek SH, Kim EK, Goudreau JL, Lookingland KJ, Kim SW, Yu SW. Insulin withdrawal-induced cell death in adult hippocampal neural stem cells as a model of autophagic cell death. Autophagy 2009;5:277-279.
- Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, Ezaki J, Mizushima N, Ohsumi Y, Uchiyama Y, Kominami E, Tanaka K, Chiba T. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol 2005;169:425-434.
- Hong CJ, Park H, Yu SW. Autophagy for the quality control of adult hippocampal neural stem cells. Brain Res 2016;1649:166-172.
- Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods 2014;11:783-784.
- Molyneaux BJ, Arlotta P, Menezes JR, Macklis JD. Neuronal subtype specification in the cerebral cortex. Nat Rev Neurosci 2007;8:427-437.
- Wang S, Li B, Qiao H, Lv X, Liang Q, Shi Z, Xia W, Ji F, Jiao J. Autophagy-related gene Atg5 is essential for astrocyte differentiation in the developing mouse cortex. EMBO Rep 2014;15:1053-1061.
- Katsuragi Y, Ichimura Y, Komatsu M. p62/SQSTM1 functions as a signaling hub and an autophagy adaptor. FEBS J 2015;282:4672-4678.
- Johansen T, Lamark T. Selective autophagy mediated by autophagic adapter proteins. Autophagy 2011;7:279-296.
- Sanz L, Diaz-Meco MT, Nakano H, Moscat J. The atypical PKC-interacting protein p62 channels NF-κB activation by the IL-1-TRAF6 pathway. EMBO J 2000;19:1576-1586.
- Folmes CD, Dzeja PP, Nelson TJ, Terzic A. Metabolic plasticity in stem cell homeostasis and differentiation. Cell Stem Cell 2012;11:596-606.
- Sin J, Andres AM, Taylor DJ, Weston T, Hiraumi Y, Stotland A, Kim BJ, Huang C, Doran KS, Gottlieb RA. Mitophagy is required for mitochondrial biogenesis and myogenic differentiation of C2C12 myoblasts. Autophagy 2016;12:369-380.
- Clarke DL, Johansson CB, Wilbertz J, Veress B, Nilsson E, Karlström H, Lendahl U, Frisén J. Generalized potential of adult neural stem cells. Science 2000;288:1660-1663.



