Articles
Article Tools
Stats or Metrics
Article
Original Article
Exp Neurobiol 2019; 28(4): 537-546
Published online August 31, 2019
https://doi.org/10.5607/en.2019.28.4.537
© The Korean Society for Brain and Neural Sciences
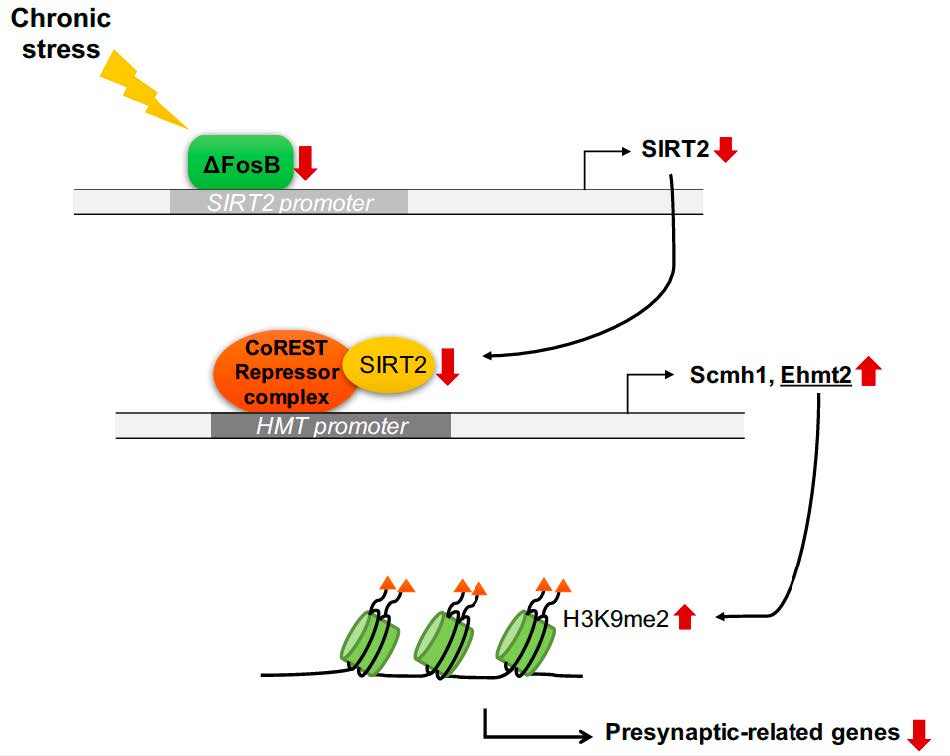
Downregulation of SIRT2 by Chronic Stress Reduces Expression of Synaptic Plasticity-related Genes through the Upregulation of Ehmt2
Sung Eun Wang1, Seung Yeon Ko1, Sungsin Jo2, Hye-Ryeong Jo1, Jinil Han3, Yong-Seok Kim1,4 and Hyeon Son1,4*
1Hanyang Biomedical Research Institute, Graduate School of Biomedical Science and Engineering, Hanyang University, Seoul 04763, 2Hanyang University Hospital for Rheumatic Diseases, Seoul 04763, 3Gencurix, Inc, Hanhwan Bizmetro 1, Seoul 08394, 4Department of Biochemistry and Molecular Biology, College of Medicine, Hanyang University, Seoul 04763, Korea
Correspondence to: *To whom correspondence should be addressed.
TEL: 82-2-2220-0626, FAX: 82-2-2220-2418
e-mail: hyeonson@hanyang.ac.kr
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License
(http://creativecommons.org/licenses/by-nc/4.0) which permits unrestricted non-commercial use, distribution, and
reproduction in any medium, provided the original work is properly cited.
Abstract
Silent information regulator 2 (Sirtuin2 / SIRT2) is a NAD+-dependent deacetylase that regulates the cellular oxidative stress response. It modulates transcriptional silencing and protein stability through deacetylation of target proteins including histones. Previous studies have shown that SIRT2 plays a role in mood disorders and hippocampus-dependent cognitive function, but the underlying neurobiological mechanism is poorly understood. Here, we report that chronic stress suppresses SIRT2 expression in the hippocampus. Molecular and biochemical analyses indicate that the stress-induced decrease in the SIRT2 expression downregulates synaptic plasticity-related genes in the hippocampus through the increase of euchromatic histone-lysine N-methyltransferase 2 (Ehmt2) (also known as G9a). shRNA-mediated knockdown of SIRT2 in the dentate gyrus alters the expression of synaptic plasticity-related genes in a way similar to those induced by chronic stress, and produces depression-like behaviors. Our results indicate that SIRT2 plays an important role in the response to stress, thereby modulating depression-like behaviors.
Graphical Abstract
Keywords: SIRT2, depression, Ehmt2, Synaptic plasticity, Hippocampus
INTRODUCTION
Silent information regulator proteins (Sirtuins / SIRTs) are nicotinamide adenine dinucleotide (NAD+)-dependent class III histone deacetylases (HDACs), which are linked to the damage occurring in neurodegenerative diseases [1]. They play key roles in mood disorders [2], and abnormal functioning of SIRT1 mediates depression-like behavior in the nucleus accumbens and hippocampus of rodents [3, 4]. However, the function of SIRT2 and its role in depression remain controversial.
Susceptibility or resilience to stress appears to be regulated by epigenetic mechanisms including histone modification [8]. Class I and II HDACs are involved in the behavioral response to chronic stress and antidepressants in rodents, suggesting the possibility of using HDAC inhibitors in patients with treatment-resistant depression [9]. Methylation of histones is an additional epigenetic mechanism involved in depression [10]. Chronic stress induces the expression of euchromatic histone-lysine N-methyltransferase 2 (Ehmt2) involved in complexes repressing transcription [11]. Ehmt2-induced di-methylated histone 3 lysine 9 (H3K9me2) is a marker of repression induced in the hippocampus and amygdala during stress and anxiety [11]. Although Ehmt2 mediates stress-induced depression-like behaviors [11], the associated transcriptional program is poorly understood.
Here we demonstrate that chronic stress-induced downregulation of SIRT2 increases expression of histone methyltransferases in the hippocampus of a mouse depression model, resulting in transcriptional repression of synaptic plasticity-related genes. Furthermore, knockdown of SIRT2 in the dentate gyrus (DG) precipitated depression-like behaviors in mice, accompanied by reduced expression of synaptic plasticity-related genes. These observations reveal a molecular mechanism that explains why hippocampal SIRT2 expression in depressive state impairs synaptic plasticity, and underline the importance of SIRT2 as a potential target for protecting synapses from depression.
MATERIALS AND METHODS
Mice
All experiments were conducted with 8–12-week-old male C57BL/6J mice (Charles River Korea, Seoul, Korea). The mice were housed 3–4 per cage in a temperature- and humidity-controlled environment under a 12-h light/dark cycle (lights on 07:00–19:00) with access to food and water ad libitum. All animals were handled in accordance with animal care guidelines of Hanyang University, and all animal experiments were approved by the Institutional Animal Care and Use Committee of Hanyang University (HY-IACUC-18-0055).
Culture of primary hippocampal neurons
Primary hippocampal neuronal were cultured as previously described [12].
Drugs
For
Chromatin immunoprecipitation assays (ChIP)
ChIP assays were performed as previously described [12]. Mouse hippocampal tissue and cultured hippocampal neurons were chemically cross-linked by the addition of formaldehyde solution and then sonicated. The resulting extract was incubated with 5 μg of the appropriate antibody and Protein G Agarose beads (Roche, Mannheim, Germany). The antibodies used are anti-ΔFosB (Cell signaling, MA, USA, #14695), anti-CoREST (Millipore, CA, USA, #07-455) and anti-H3K9me2 (Cell signaling, MA, USA, #4658). Immunoprecipitated DNA samples were resuspended in distilled H2O and used in real-time PCR (CFX96 TouchTM Real-Time PCR Detection System, Bio-Rad Laboratories, CA, USA), using the following primer pairs corresponding to mouse gene promoter regions:
Immunohistochemistry and Western blot analysis
Mice were perfused with 4% paraformaldehyde in PBS for 20 min and processed for histology. Immunofluorescence labeling and Western blot analysis were performed as previously described [12]. The primary antibodies used for immunohistochemistry and Western blot analysis were as follows: β-actin (Santa Cruz, sc-47778), ΔFosB (Cell signaling, #14695), SIRT2 (Millipore, 09-843), Egr1 (Cell signaling, #4153), Tuj1 (Covance, MMS-435P), GluR1 (Upstate, # 06-306), GluR2 (Millipore, AB1768-I), NR1 (Upstate, #06-311), PSD95 (abcam, ab18258), Synaptophysin (abcam, ab32127), Synapsin1 (Cell signaling, #5297), CoREST (Millipore, #07-455), and GFP (Roche Applied Sciences, 11 814 460 001).
Co-immunoprecipitation
Hippocampal neuron lysates were generated by using lysis buffer (Cell Signaling, MA, USA) contained protease inhibitor and phosphatase inhibitor I and II (Sigma). 2 μg antibody (CoREST, Millipore, CA, USA, #07-455) or rabbit IgG (Millipore, CA, USA) was added to lysates containing 500 μg protein at 4°C overnight [12], which was followed by immunoprecipitation with 30 μl Protein G Agarose beads (Roche, Mannheim, Germany). Beads were washed five times with HNTG buffer (20 mM HEPES pH 7.5, 150 mM NaCl, 0.1% Triton X-100 and 10% glycerol). The bound proteins were eluted with 2X Laemmli sample buffer (126 mM Tris-HCl, 20% glycerol, 4% SDS, 0.02% bromophenol blue) and analyzed by Western blotting.
RT-PCR and qPCR
RNA was extracted from hippocampi with Trizol reagent (Sigma), and reverse transcribed with Improm-II (Promega, WI, USA), 1 μg of total RNA and oligonucleotide-dT primer. Quantitative real-time PCR (qPCR) was performed on a CFX96 TouchTM Real-Time PCR Detection System (Bio-Rad Laboratories) using the primer sets of 5′-AGCCAACCATCTGCCACTAC-3′ and 5′-CCAGCCCATCGTGTA TTCTT-3′ for
Construction of a short hairpin RNA (shRNA)-expressing vector, and lentivirus production
For
Virus-mediated gene transfer
Mice were anesthetized with a mixture of ketamine (100 mg/ kg) and xylazine (10 mg/kg) and prepared for stereotaxic surgery. Thirty-three gauge syringe needles (Hamilton, NV, USA) were used to bilaterally infuse 3 μl of virus into the dorsal hippocampus at a rate of 0.1 μl/min at the stereotaxic coordinates −2.0 anteroposterior (AP), ±1.5 mediolateral (ML), −2.4 dorsoventral (DV) (mm from bregma, horizontal skull).
Chronic stress procedures
Chronic unpredictable stress (CUS) experiments were performed as described previously. Mice were exposed to a sequence of mild and unpredictable stressors for 14 d [12]. The 11 stressors used to generate the animal model of depression were as previously described [15]. For the chronic restraint stress (CRS) procedure, the mice were restrained horizontally in 50 ml conical centrifuge tubes drilled with holes to allow ample ventilation; the tubes were small enough to restrain the mice so that they were able to breathe but unable to move freely. The mice were stressed daily for 2 h for 14 d, and provided with food and water ad libitum [16].
Assessment of behavior
On the test day, mice were placed to the testing room and acclimated to the room conditions for at least 1 h. Mice of the same cage were randomly assigned to different experimental groups for behavioral studies, and the order of testing was distributed across groups. All tests were conducted during the dark cycle. After each individual test session, the apparatus was thoroughly cleaned with 70% alcohol to eliminate any odor and trace of the previously tested mouse. Wherever possible, the experimenter conducting the data analysis was blind to the treatment conditions of the animals.
Locomotor activity test (LMA)
Total locomotor activity was measured for 5 min with an ANY-maze video tracking system (Stoelting Co., IL, USA) in a plastic box (50×50×20 cm) and conducted as previously described [12].
Forced swim test (FST)
FST was conducted as previously described. Mice were placed in a cylinder (height: 30 cm, diameter: 15 cm) filled with water at 24±1°C and a depth of 12 cm for 6 min. The immobility time in the last 4 min of the 6 min test period were measured [12].
Learned helplessness test (LHT)
The learned helplessness procedure was based on previously described procedures. Mice were placed on a commercial shuttle box divided into two equal compartments by a central barrier (Gemini Avoidance System, San Diego Instruments) and then given inescapable footshocks (0.3 mA, 180 scrambled footshocks). The mean escape latency over 30 successive escape trials (0.3 mA, 25-sec maximum duration) was measured [12, 17].
Microarray dataset analysis
The following analyzing method was previously described [18]. The gene expression microarray dataset was obtained from the Gene Expression Omnibus (GEO,
Statistical analyses
Data distributions were checked for normality and equal variance. All experiments were carried out at least three times, and data consistency was observed in repeated experiments. GraphPad Prism5.0 (GraphPad Software, Inc., La Jolla, CA, USA) was used for statistical analysis. p<0.05 was considered statistically significant. Biochemical data are presented as means±s.e.m. of multiple independent experiments performed in triplicate.
RESULTS
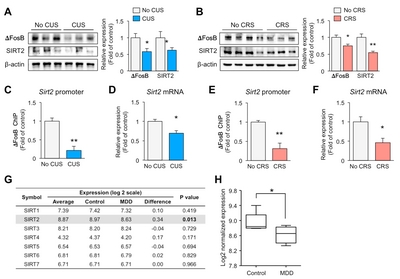
Chronic stress downregulates exprssions of ΔFosB and SIRT2 in the hippocampus of mice
SIRT2 expression is decreased in the blood of depressive-state MDD patients, and restored in the remissive state [5]. To examine the role of SIRT2 in the hippocampus following exposure to stress, we obtained mice that had been exposed to CUS or CRS for 2 weeks. We confirmed the reduced levels of SIRT2 in the hippocampus of the CUS (Fig. 1A) and CRS (Fig. 1B) models. Expression of delta FosB (ΔFosB), a transcription factor that is involved in stress [21] and known to directly regulate SIRT2 expression [22], was reduced in both depression models (Fig. 1A and 1B). To investigate direct regulation by ΔFosB of the
Expression of SIRT2 is decreased in the hippocampus in MDD
To see if our results could be translated to human, we evaluated
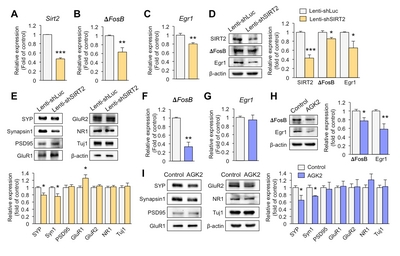
Downregulation of SIRT2 reduces the expression of synaptic plasticity-related genes in primary hippocampal neurons
To investigate whether knockdown of SIRT2 affects expression of synaptic plasticity-related genes, we introduced lenti-shSIRT2 into hippocampal primary neurons to reduce SIRT2 expression. SIRT2 expression was markedly attenuated at both mRNA and protein levels (Fig. 2A and 2D). ΔFosB is known to directly regulate SIRT2 expression but we also found that SIRT2 knockdown decreased expression of ΔFosB (Fig. 2B and 2D). Early growth response protein 1 (Egr1) is thought to play a key role in inducing synaptic plasticity-related genes [23], and SIRT2 knockdown reduced the expression of Egr1 together with the pre-synaptic molecules, Synaptophysin and Synapsin1 (Fig. 2C~E). In addition, expression of glutamate receptor 1 (GluR1) was increased in lenti-shSIRT2 infected neurons compared to lenti-shLuc infected neurons (Fig. 2E), consistent with an elevated level of GluR1 as a result of its increased stability due to reduced deacetylation by SIRT2 [24]. To see if a SIRT2 inhibition had similar effects to SIRT2 knockdown, we treated primary hippocampal neurons with AGK2 (5 μM, 72 h), an inhibitor of SIRT2, and found that expression levels of ΔFosB and pre-synaptic molecules including Synaptophysin and Synapsin1 were decreased, while that of Egr1 did not (Fig. 2F~I). Together, these results indicate that SIRT2 regulates the expression of transcription factors and pre-synaptic molecules that play important roles in resilience to stress.
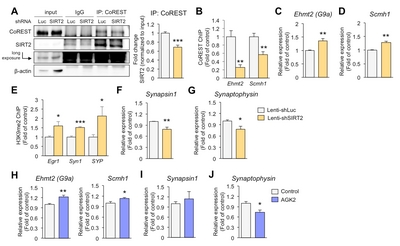
SIRT2-mediated suppression of Ehmt2 increases presynaptic gene expression in hippocampal neurons
It is well known that SIRT2 targets lysines in various cytosolic proteins [25]. However, we have focused on the function of SIRT2 in the nucleus, where transcriptional regulation occurs. SIRT2 is recruited to specific loci by co-repressor element-1 silencing transcription factor (CoREST) [26]. To study whether the inhibition of synaptic plasticity-related genes is regulated by SIRT2 knockdown, we first examined the association of SIRT2 with its corepressor and observed decreased association in primary hippocampal neurons infected with lenti-shSIRT2 compared to neurons infected with lenti-shLuc (Fig. 3A). Given that SIRT2-CoREST corepressor complexes would inhibit the transcription of genes [26], we would expect that expression of SIRT2 target genes was increased in lenti-shSIRT2 infected neurons. We found that coimmunoprecipitation of SIRT2 and CoREST was reduced in neurons that were infected by lenti-shSIRT2 compared to neurons infected with lenti-shLuc (Fig. 3A). CoREST binding to the promoter region of Ehmt2 and Scm Polycomb Group Protein Homolog 1 (Scmh1) was decreased in neurons infected by lenti-shSIRT2 (Fig. 3B). Consistently, mRNA levels of Ehmt2 and Scmh1, two histone methyltransferases that targeted by SIRT2, were increased in primary hippocampal neurons infected with lenti-shSIRT2 compared to neurons infected with lenti-shLuc (Fig. 3C and 3D), suggesting that SIRT2 negatively regulates the expression of Ehmt2 and Scmh1 although it is not clear whether this is a direct or indirect effect. Furthermore, H3K9me2, a known target of Ehmt2 and a marker of repressed gene repression, was enriched on the promoter regions of
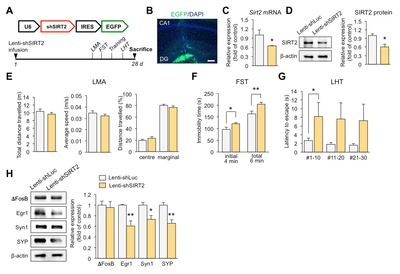
Knockdown of SIRT2 in the DG induces depression-like behaviors in mice
To demonstrate behavioral effects of SIRT2 knockdown, we infused lenti-shSIRT2 bilaterally into the hippocampal DG, which region is vulnerable to stress [27] (Fig. 4A and 4B). SIRT2 expression was reduced at both the mRNA and protein level (Fig. 4C and 4D). We then evaluated whether SIRT2 knockdown increased stress-induced behaviors, as indicated by reduced responsiveness in the FST and the LHT, two widely used models of behavioral despair. We found that immobility time and the latency to escape increased in the lenti-shSIRT2-infused mice (Fig. 4F and 4G). There was no effect on spontaneous locomotor activity, including average speed, total distance travelled, and distance travelled from the center or margin, indicating that the effects in the FST were not due simply to inhibition of mobility (Fig. 4E). Consistent with the
DISCUSSION
This study demonstrates that CUS downregulates SIRT2, resulting in an increase in the Ehmt2 which functions in the CoREST repressor complex. Ehmt2 targets the promoters of synaptic plasticity-related genes such as
Here, we show that H3K9me2, a marker of repressive genes, accumulates in the promoter regions of
According to previous reports, SIRT2 has a major role in modulating diverse biological mechanisms in the cytosol, including oxidative stress and inflammatory response in microglia cells associated with CNS disorders [2]. SIRT2 downregulation leads to cognitive impairment through aberrant post-translational modification of the AMPA receptor (AMPAR) in the CNS, which is directly related to depressive states [24, 31]. The function of SIRT2 in the nucleus is poorly understood; it appears to be a kind of histone deacetylase but its underlying role in depression is unclear. To the best of our knowledge, our findings constitute the first report that downregulation of SIRT2 produces depression-like behaviors via increased expression of histone methyltransferase that represses the promoter regions of synaptic plasticity-related genes in the hippocampus, and therefore, SIRT2 may be an attractive therapeutic target in major depressive disorder.
ACKNOWLEDGEMENTS
This work was supported by a National Research Foundation of Korea (NRF) grant (No. 2019R1A2C2003616 to H.S.), a Medical Research Center Grant (2017R1A5A2015395 to H.S.) and a Basic Science Research Program Grant (No. 2017R1A6A3A01005765 to S.E.W.) funded by the Ministry of Science and Technology (MEST), Republic of Korea.
Figures
References
- Min SW, Sohn PD, Cho SH, Swanson RA, Gan L (2013) Sirtuins in neurodegenerative diseases: an update on potential mechanisms. Front Aging Neurosci 5 : 53.
- Song J, Kim J (2016) Role of sirtuins in linking metabolic syndrome with depression. Front Cell Neurosci 10 : 86.
- Kim HD, Hesterman J, Call T, Magazu S, Keeley E, Armenta K, Kronman H, Neve RL, Nestler EJ, Ferguson D (2016) SIRT1 mediates depression-like behaviors in the nucleus accumbens. J Neurosci 36 : 8441-8452.
- Abe-Higuchi N, Uchida S, Yamagata H, Higuchi F, Hobara T, Hara K, Kobayashi A, Watanabe Y (2016) Hippocampal sirtuin 1 signaling mediates depression-like behavior. Biol Psychiatry 80 : 815-826.
- Abe N, Uchida S, Otsuki K, Hobara T, Yamagata H, Higuchi F, Shibata T, Watanabe Y (2011) Altered sirtuin deacetylase gene expression in patients with a mood disorder. J Psychiatr Res 45 : 1106-1112.
- Liu R, Dang W, Du Y, Zhou Q, Jiao K, Liu Z (2015) SIRT2 is involved in the modulation of depressive behaviors. Sci Rep 5 : 8415.
- Erburu M, Muñoz-Cobo I, Domínguez-Andrés J, Beltran E, Suzuki T, Mai A, Valente S, Puerta E, Tordera RM (2015) Chronic stress and antidepressant induced changes in Hdac5 and Sirt2 affect synaptic plasticity. Eur Neuropsychopharmacol 25 : 2036-2048.
- Tsankova N, Renthal W, Kumar A, Nestler EJ (2007) Epigenetic regulation in psychiatric disorders. Nat Rev Neurosci 8 : 355-367.
- Covington HE 3rd, Maze I, LaPlant QC, Vialou VF, Ohnishi YN, Berton O, Fass DM, Renthal W, Rush AJ 3rd, Wu EY, Ghose S, Krishnan V, Russo SJ, Tamminga C, Haggarty SJ, Nestler EJ (2009) Antidepressant actions of histone deacetylase inhibitors. J Neurosci 29 : 11451-11460.
- Sun H, Kennedy PJ, Nestler EJ (2013) Epigenetics of the depressed brain: role of histone acetylation and methylation. Neuropsychopharmacology 38 : 124-137.
- Covington HE 3rd, Maze I, Sun H, Bomze HM, DeMaio KD, Wu EY, Dietz DM, Lobo MK, Ghose S, Mouzon E, Neve RL, Tamminga CA, Nestler EJ (2011) A role for repressive histone methylation in cocaine-induced vulnerability to stress. Neuron 71 : 656-670.
- Wang SE, Ko SY, Jo S, Choi M, Lee SH, Jo HR, Seo JY, Lee SH, Kim YS, Jung SJ, Son H (2017) TRPV1 regulates stress responses through HDAC2. Cell Reports 19 : 401-412.
- Narayan N, Lee IH, Borenstein R, Sun J, Wong R, Tong G, Fergusson MM, Liu J, Rovira II, Cheng HL, Wang G, Gucek M, Lombard D, Alt FW, Sack MN, Murphy E, Cao L, Finkel T (2012) The NAD-dependent deacetylase SIRT2 is required for programmed necrosis. Nature 492 : 199-204.
- Kwon M, Firestein BL (2013) DNA transfection: calcium phosphate method. Methods Mol Biol 1018 : 107-110.
- Schmidt HD, Duman RS (2010) Peripheral BDNF produces antidepressant-like effects in cellular and behavioral models. Neuropsychopharmacology 35 : 2378-2391.
- Kim KS, Han PL (2006) Optimization of chronic stress paradigms using anxiety- and depression-like behavioral parameters. J Neurosci Res 83 : 497-507.
- Duman CH, Schlesinger L, Kodama M, Russell DS, Duman RS (2007) A role for MAP kinase signaling in behavioral models of depression and antidepressant treatment. Biol Psychiatry 61 : 661-670.
- Ko SY, Wang SE, Lee HK, Jo S, Han J, Lee SH, Choi M, Jo HR, Seo JY, Jung SJ, Son H (2019) Transient receptor potential melastatin 2 governs stress-induced depressive-like behaviors. Proc Natl Acad Sci U S A 116 : 1770-1775.
- Lanz TA, Joshi JJ, Reinhart V, Johnson K, Grantham LE 2nd, Volfson D (2015) STEP levels are unchanged in pre-frontal cortex and associative striatum in post-mortem human brain samples from subjects with schizophrenia, bipolar disorder and major depressive disorder. PLoS One 10 : e0121744.
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK (2015) limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43 : e47.
- Nestler EJ (2015) ΔFosB: a transcriptional regulator of stress and antidepressant responses. Eur J Pharmacol 753 : 66-72.
- Renthal W, Kumar A, Xiao G, Wilkinson M, Covington HE 3rd, Maze I, Sikder D, Robison AJ, LaPlant Q, Dietz DM, Russo SJ, Vialou V, Chakravarty S, Kodadek TJ, Stack A, Kabbaj M, Nestler EJ (2009) Genome-wide analysis of chromatin regulation by cocaine reveals a role for sirtuins. Neuron 62 : 335-348.
- Duclot F, Kabbaj M (2017) The role of early growth response 1 (EGR1) in brain plasticity and neuropsychiatric disorders. Front Behav Neurosci 11 : 35.
- Wang G, Li S, Gilbert J, Gritton HJ, Wang Z, Li Z, Han X, Selkoe DJ, Man HY (2017) Crucial roles for SIRT2 and AMPA receptor acetylation in synaptic plasticity and memory. Cell Reports 20 : 1335-1347.
- Kazantsev AG, Thompson LM (2008) Therapeutic application of histone deacetylase inhibitors for central nervous system disorders. Nat Rev Drug Discov 7 : 854-868.
- Cadet JL, Jayanthi S (2013) Epigenetics of methamphetamine-induced changes in glutamate function. Neuropsychopharmacology 38 : 248-249.
- McEwen BS, Nasca C, Gray JD (2016) Stress effects on neuronal structure: hippocampus, amygdala, and prefrontal cortex. Neuropsychopharmacology 41 : 3-23.
- Thiel G, Schoch S, Petersohn D (1994) Regulation of synapsin I gene expression by the zinc finger transcription factor zif268/egr-1. J Biol Chem 269 : 15294-15301.
- Shi YJ, Matson C, Lan F, Iwase S, Baba T, Shi Y (2005) Regulation of LSD1 histone demethylase activity by its associated factors. Mol Cell 19 : 857-864.
- Wang DY, Kosowan J, Samsom J, Leung L, Zhang KL, Li YX, Xiong Y, Jin J, Petronis A, Oh G, Wong AH (2018) Inhibition of the G9a/GLP histone methyltransferase complex modulates anxiety-related behavior in mice. Acta Pharmacol Sin 39 : 866-874.
- Kallarackal AJ, Kvarta MD, Cammarata E, Jaberi L, Cai X, Bailey AM, Thompson SM (2013) Chronic stress induces a selective decrease in AMPA receptor-mediated synaptic excitation at hippocampal temporoammonic-CA1 synapses. J Neurosci 33 : 15669-15674.





