Articles
Article Tools
Stats or Metrics
Article
Original Article
Exp Neurobiol 2021; 30(3): 203-212
Published online June 30, 2021
https://doi.org/10.5607/en21017
© The Korean Society for Brain and Neural Sciences
Cell Type-specific Knockout with Gli1 -mediated Cre Recombination in the Developing Cerebellum
Jung-Mi Choi1†, Rakshya Acharya1,2†, Subash Marasini3, Bashyal Narayan1,2, Kwang-Wook Lee1, Woo Sup Hwang1, Da-Young Chang3, Sung-Soo Kim1,2* and Haeyoung Suh-Kim1,2,3*
1Department of Anatomy, Ajou University School of Medicine, Suwon 16499, 2Department of Biomedical Sciences, Graduate School, Ajou University School of Medicine, Suwon 16499, 3Research Center, CelleBrain Ltd., Jeonju 54871, Korea
Correspondence to: *To whom correspondence should be addressed.
Sung-Soo Kim, TEL: 82-31-219-5036, FAX: 82-31-219-5034
e-mail: kimdmg@ajou.ac.kr
Haeyoung Suh-Kim, TEL: 82-31-219-5036, FAX: 82-31-219-5039
e-mail: hysuh@ajou.ac.kr
†These authors contributed equally to this work.
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/4.0) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Abstract
The inducible
Graphical Abstract
Keywords: Cerebellum, Cre recombinase, Tamoxifen, Gli1, Bergmann glia, Granule cell neuron
INTRODUCTION
The Cre-loxP recombination system has been widely used for studying gene functions in animals by allowing region-specific knockout of target genes through site-specific expression of Cre. Inducible Cre such as CreER provides more specific control of spatiotemporal deletion or lineage labeling through timed administration of synthetic estrogen receptor (ER) ligands such as tamoxifen (TAM) or 4-hydroxytamoxifen (4-OHT) [1, 2]. Embryonic administration of tamoxifen rapidly induces abortion in pregnant mouse mothers and severely perturbs embryonic development, rendering the inducible Cre system inapplicable to the study of developmentally regulated genes in embryos [3]. In contrast, tamoxifen injection after birth is less harmful and relatively tolerable in neonates, suggesting that inducible Cre-loxP may provide genetic tools for the study of postnatal development [4]. While most major structures in the central nervous system develop before birth, cerebellar architecture develops actively during the first three weeks after birth [5]. During this period, neuronal progenitor cells proliferate, migrate, and terminally differentiate into the cerebellar cortex. Postnatal injection of tamoxifen to neonates may enable studies on developmentally regulated gene functions in mitotic and/or postmitotic cells in the cerebellum.
The cerebellar cortex consists of three distinct layers: the molecular layer (ML), Purkinje cell layer (PCL), and granule cell layer (GCL). The somata of Purkinje cells (PCs) and Bergmann glia (BGs) are arranged in a single PCL layer. PCs are the only output neurons of the cerebellar cortex, and each sends a single, long axon to the deep cerebellar nuclei (DCN). ML contains inhibitory interneurons, parallel fibers of GCNs, PC dendrites, and BG radial fibers. The complex dendrites of PCs in the ML receive presynaptic inputs from parallel fibers (PFs) originating in GCNs in the GCL and climbing fibers (CFs) projecting from the inferior olivary nucleus [6]. This trilaminar architecture of the cerebellar cortex developed perinatally. During the late embryonic period (E17.5) and postnatal development, cerebellar granule cell progenitors (GCPs) rapidly proliferate in the external granule layer (EGL), radially migrate to the internal GCL, and eventually differentiate into GCNs. Sonic hedgehog (Shh) plays a key role in the proliferation of GCPs during cerebellar morphogenesis and histogenesis, and deletion of Shh induces hypoplasia of the cerebellar cortex [7-9]. Shh secreted by PC [9, 10] activates the Gli1 promoter in GCPs and BGs [7, 11-13]. Thus,
In this study, we investigated whether
MATERIALS AND METHODS
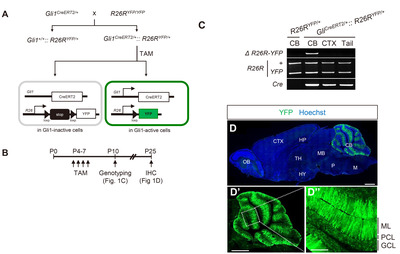
Mouse genetics
TAM (Sigma-Aldrich) was dissolved in corn oil to a final concentration of 10 mg/ml. To activate Cre recombinase, mice were force-fed TAM (50 μg/g weight/ day) by oral pipet-feeding at the indicated time points and housed until euthanized.
All experimental procedures were approved by Ajou University Medical Center-Institutional Animal Care and Use Committee (AUMC-IACUC, Suwon, South Korea).
Immunofluorescence analysis
Immunohistochemical analyses were performed as described previously [20]. Briefly, mice were deeply anesthetized with 2,2,2 tribromoethanol (200 mg/kg, i.p., Sigma-Aldrich), and then perfused transcardially with 10% neutral buffered formalin (BBC Biochemical). The brain was extracted, post-fixed in 10% neutral buffered formalin overnight at 4°C, and cryoprotected in 30% sucrose. The brain was sagitally divided into two halves, embedded in OCT compound (Tissue-Tek, Sakura Finetek), and sectioned into 30 μm-thick frozen sections using a Leica cryostat (Leica). The cryosections were air-dried, and the residual OCT compound was washed in PBS with 0.1% (v/v) Triton X-100 (Sigma-Aldrich) (PBS-T). After incubation in blocking solution [10% (vol/vol) normal goat serum (Gibco), 1% bovine serum albumin (BSA; Sigma-Aldrich) in PBS-T] for 1 h at room temperature, the sections were incubated with primary antibodies overnight at 4°C. The antibodies used in this study were as follows: anti-GFP (1:500, Abcam, #ab13970), anti-Pcp2 (1:500, Santa Cruz, #sc-137064), anti-GFAP (1:200, Dako, #Z0334), anti-S100β (1:500, Abcam, #ab41548), anti-NeuN (1:500, EMD Millipore, #MAB377), and anti-parvalbumin (PV; 1:500; Swant, #PV25). After unbound antibodies were washed with PBS-T, the sections were incubated with secondary antibodies conjugated with Alexa Fluor 405, 488, or 568 (1:500, Invitrogen). If necessary, nuclear counterstaining was performed using bisbenzamide (1:50,000, Hoechst 33258; Invitrogen). All fluorescence images were acquired using a Zeiss LSM710 confocal laser scanning microscope (Carl Zeiss) or Zeiss Axio Scan Z1 slide scanner (Carl Zeiss) at the Three-Dimensional Immune System Imaging Core Facility of Ajou University.
Quantitative analysis
Confocal images of cerebellar sagittal sections were analyzed using ZEN software (Blue Edition, Zeiss). The specificity of
RESULTS
Cerebellum specific Cre-mediated recombination driven by Gli1 promoter
To assess the Gli1 promoter-mediated expression of the Cre enzyme,
The TAM-activated Cre enzyme excised the stop cassette flanked by two loxP sites and permitted YFP expression in Gli1-expressing cells. Cre-mediated recombination was validated using a 750 bp PCR product with gDNA isolated from the cerebellum. Such PCR products were not detected in the cerebral cortex, suggesting that the Gli1 promoter is active only in the cerebellum (Fig. 1C). Consistently, in the sagittal sections of
Gli1 active cells in early postnatal cerebellum
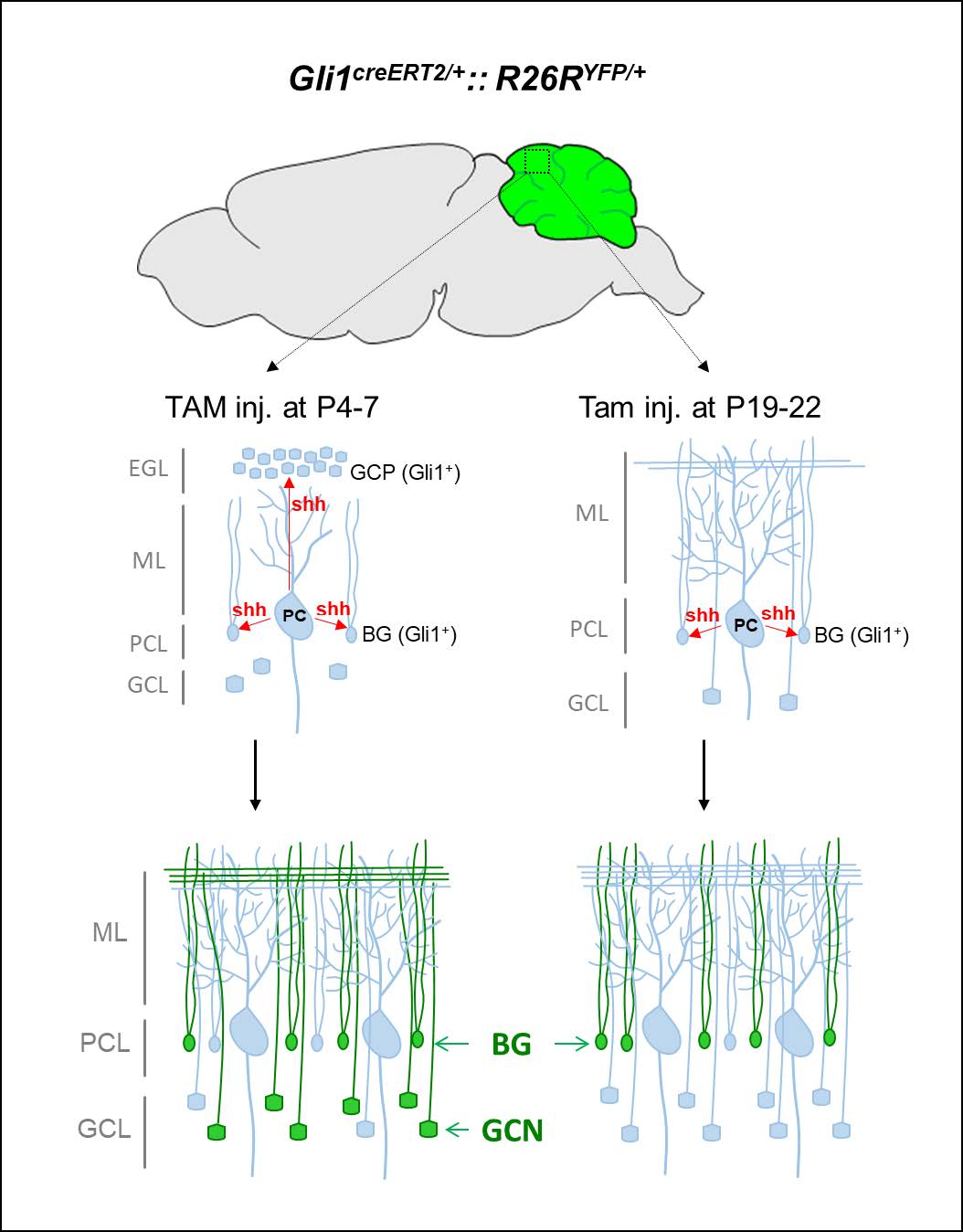
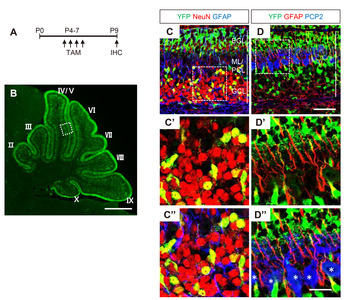
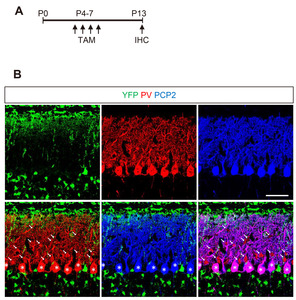
The proliferation of GCPs reaches its peak at P4-8 in response to Shh produced by PCs [22]. To determine the cell types responsive to Shh in early postnatal period, we administered TAM at the peak time of the GCP proliferating period (P4~7) and sacrificed the mice at P9 (Fig. 2A). YFP expression was detected in proliferating GPCs in EGL. YFP+ cells migrated to the GCL and became fully differentiated NeuN+ GCNs. YFP expression was also detected in radially extending fibers in the ML and soma of GFAP+ BGs in the PCL (Fig. 2C and 2D). YFP was not expressed in Pcp2+ PCs in the PCL (asterisks in Fig. 2D”) or parvalbumin (PV)+ including GABAergic inerneurons and PCs (Fig. 3). These results suggest that the administration of tamoxifen during early postnatal days induces expression of Cre recombinase in Gli1-expression GCPs and BGs, but not in other types of cells in the developing cerebellum.
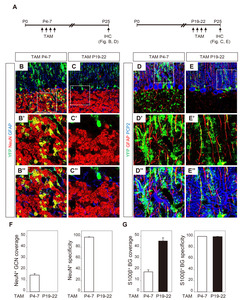
Cerebellar granule cells- and Bergmann glial cells- specific expression of Cre recombinase activity
To determine the duration of Shh-responsiveness, we administered TAM at two distinct time points: during P4~7, when GCPs in the EGL migrated to the internal GCL and during P19~22 when EGL no longer existed [23]. YFP expression that was induced by TAM at P4~7 in GCPs and BGs was maintained in GCNs and BGs in the mature brain at P25 (Fig. 4B and 4D). In contrast, TAM administration at P19~22 induced YFP expression only in BGs, but not in GCs (Fig. 4C and 4E). For quantification, we performed additional staining with anti-S100β. GFAP and S100β revealed the same cell population with distinctive immunoreactivity: S100β in the somata and proximal processes of BGs and GFAP in the arborized glial fibers. TAM injection at P4~7 induced YFP+ expression in 15.1±1.4% of NeuN+ GCNs and 18.0±1.8% of BGs (Fig. 4F and 4G). The specificity of YFP expression was high thus most YFP+ cells in GCL or PCL were NeuN+ GCN (98.6±0.46%) or BGs (100%), respectively. When TAM was injected at P19~22, the YFP+ expression was found in 45.8±2.9% to BGs in PCL with 99.5±0.47% specificity. None of GCNs were co-localized with YFP expression. The results indicated that Shh signaling is temporarily active in the proliferation of GCP and BG during the early postnatal period and constitutively active only in BGs.
DISCUSSION
This study showed that timed postnatal administration of TAM differentially regulates cell-type-specific excision of floxed genes in the developing cerebellar cortex. TAM administration at early postnatal days (P4~7), when the GCP proliferation peaks in the EGL, can induce the expression of Gli1-mediated Cre recombinase in GCPs of EGL and BGs in PCL. Thus, tamoxifen administration during postnatal days (P19~22), when EGL is almost depleted, leads to the expression of YFP only in BGs. Our results are consistent with the previous finding that Gli1 expression is restricted to proliferating GCPs and BGs in the developing cerebellar cortex in response to Purkinje-derived Shh in postnatal stages through adulthood [10, 13, 24, 25]. Importantly, YFP+ GCPs in the EGL inwardly migrate to and differentiate NeuN+ GCNs in the GCL, where they remain as YFP+ GCNs to adulthood (Fig. 2). As mentioned earlier, proteins with long half-lives in the synaptic compartment [17] may remain in postmitotic GCNs even after the mRNA and protein are no longer synthesized.
Shh-Gli1 signaling in the developing cerebellum
All cerebellar neurons are generated from progenitors in two distinct germinative centers in the hindbrain: the rhombic lip and the ventricular zone [5, 26]. The progenitor cells in the rhombic lip express Math-1 (mouse homolog-1 of Drosophila Atonal) and generate glutamatergic neurons, including projection neurons in deep cerebellar nuclei, unipolar brush cells, and GCNs in the GCL [27, 28]. The progenitor cells in the ventricular zone generate all GABAergic phenotypes, including PCs, nucleo-olivary projection neurons, and all inhibitory interneurons, astrocytes, and oligodendrocytes in the white matter [29, 30]. Shh produced by PCs acts as a mitogen on progenitor cells originating from the rhombic lip and ventricular zone [24, 31] affects Bergmann glial differentiation [12]. Based on Gli1 expression, a high level of positive Shh signaling is restricted to the proliferating GCPs and BGs in developing cerebellar cortex [13, 24]. Only BGs have been shown to be capable of responding to PC-derived Shh signals in postnatal stages through adulthood [25]. Consistently, TAM activates Gli1-
Cre-mediated knockout for the study of cerebellar cortex
Several Cre lines have been used to knock out genes in specific cell types in the cerebellum (Table 2):
The perspective application of Gli1-CreERT2
The cerebellum is the largest sensorimotor structure in the brain and has extensive connections with the brainstem and spinal cord. The cerebellum plays an important role in coordinating skilled voluntary movements by influencing muscle activity and controlling equilibrium and muscle tone through connections with the vestibular system and the spinal cord and its gamma motor neurons. Recently, there has been rapidly increasing evidence indicating the role of the cerebellum in emotion and cognition in addition to movement [49, 50]. Long-term depression (LTD) is considered a cellular mechanism for cerebellar motor learning and is expressed as reduced responsiveness to transmitter glutamate [51, 52]. In particular, PF-PC synapses are well-known sites for LTD [53]. Intensive studies using Pcp2/L7-Cre have revealed that the molecular machinery, including Ca2+ influx, protein kinase C, and endocytosis of AMPA-type glutamate receptors play critical roles in postsynaptic PCs [41]. By comparison, the presynaptic roles of PF in LTD induction are relatively unknown.
We show that
ACKNOWLEDGEMENTS
We thank Prof. Mi-Ryoung Song (Gwangju Institute of Science and Technology) and Prof. Yongsu Jeong (Kyung Hee University) for their informative and kind advice on mouse genetics. This research was supported by grants from the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare (HI20C0457 to HS-K), the Ministry of Food and Drug Safety in 2021 (18172MFDS182-5 to HS-K), and the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF-2020R1I1A1A01053066 to J-MC) of the Republic of Korea.
Figures
Tables
The primer sequences for PCR reaction
| Target | Primer sequence | PCR product |
|---|---|---|
| Forward | 5’-AAA GTC GCT CTG AGT TGT TAT-3’ | WT: 600 bp (F+R1) Knock-in: 325 bp (F+R2) |
| Reverse 1 | 5’-GGA GCG GGA GAA ATG GAT ATG-3’ | |
| Reverse 2 | 5’-GCG AAG AGT TTG TCC TCA ACC-3’ | |
| Forward | 5’-GCG AAG AGT TTG TCC TCA ACC-3’ | 750 bp |
| Reverse | 5’-ATG GCG GAC TTG AAG AAG TCG TG-3’ | |
| Forward | 5’-GCA TTA CCG GTC GAT GCA ACG AGT GAT GAG-3’ | 404 bp |
| Reverse | 5’-GAG TAG ACG AAC CTG GTC GAA ATC AGT GCG-3’ |
Mouse lines expressing Cre recombinase in the cerebellum
| Mouse line | TAM @ | GCN | BG | PC | IN | References |
|---|---|---|---|---|---|---|
| P1~3 | V | V* | SC, BC | [15] | ||
| P4~7 | V | V | This study | |||
| Adult | V | This study, [16] | ||||
| NSE-CreERT2 | P1~3 | V | [42] | |||
| Adult | V | [42] | ||||
| P1~3 | V | UBC | [15] | |||
| P4~7 | V* | [45] | ||||
| Adult | V | [43] | ||||
| P1~3 | V* | SC, BC, UBC | [44] | |||
| P4~7 | V* | [44] | ||||
| Adult | V | [46, 47] | ||||
| P1~3 | V * | SC, BC | [15] | |||
| NA | V | [35, 36] | ||||
| NA | V | UBC | [37, 38] | |||
| NA | V | V* | [48] | |||
| NA | V | [39, 40] | ||||
| Shh-Cre | NA | V | [15] | |||
GCN, granule cell neuron; BG, Bergman glia; PC, Purkinje cell; IN, interneuron; *, cerebellar astrocyte; SC, stellate cells; BC, basket cell; UBC, Unipolar brush cells located in GCL; NA, not applicable.
References
- Kühn R, Schwenk F, Aguet M, Rajewsky K (1995) Inducible gene targeting in mice. Science 269:1427-1429
- Vaillant C, Monard D (2009) SHH pathway and cerebellar development. Cerebellum 8:291-301
- Ved N, Curran A, Ashcroft FM, Sparrow DB (2019) Tamoxifen administration in pregnant mice can be deleterious to both mother and embryo. Lab Anim 53:630-633
- Lizen B, Claus M, Jeannotte L, Rijli FM, Gofflot F (2015) Perinatal induction of Cre recombination with tamoxifen. Transgenic Res 24:1065-1077
- Carletti B, Rossi F (2008) Neurogenesis in the cerebellum. Neuroscientist 14:91-100
- Kitazawa S, Wolpert DM (2005) Rhythmicity, randomness and synchrony in climbing fiber signals. Trends Neurosci 28:611-619
- Wechsler-Reya RJ, Scott MP (1999) Control of neuronal precursor proliferation in the cerebellum by Sonic hedgehog. Neuron 22:103-114
- Fuccillo M, Joyner AL, Fishell G (2006) Morphogen to mitogen: the multiple roles of hedgehog signalling in vertebrate neural development. Nat Rev Neurosci 7:772-783
- De Luca A, Cerrato V, Fucà E, Parmigiani E, Buffo A, Leto K (2016) Sonic hedgehog patterning during cerebellar development. Cell Mol Life Sci 73:291-303
- Lewis PM, Gritli-Linde A, Smeyne R, Kottmann A, McMahon AP (2004) Sonic hedgehog signaling is required for expansion of granule neuron precursors and patterning of the mouse cerebellum. Dev Biol 270:393-410
- Wallace VA (1999) Purkinje-cell-derived Sonic hedgehog regulates granule neuron precursor cell proliferation in the developing mouse cerebellum. Curr Biol 9:445-448
- Dahmane N; Ruiz I Altaba A (1999) Sonic hedgehog regulates the growth and patterning of the cerebellum. Development 126:3089-3100
- Corrales JD, Rocco GL, Blaess S, Guo Q, Joyner AL (2004) Spatial pattern of Sonic hedgehog signaling through Gli genes during cerebellum development. Development 131:5581-5590
- Ahn S, Joyner AL (2004) Dynamic changes in the response of cells to positive hedgehog signaling during mouse limb patterning. Cell 118:505-516
- Fleming JT, He W, Hao C, Ketova T, Pan FC, Wright CC, Litingtung Y, Chiang C (2013) The Purkinje neuron acts as a central regulator of spatially and functionally distinct cerebellar precursors. Dev Cell 27:278-292
- Ye L, Orynbayev M, Zhu X, Lim EY, Dereddi RR, Agarwal A, Bergles DE, Bhat MA, Paukert M (2020) Ethanol abolishes vigilance-dependent astroglia network activation in mice by inhibiting norepinephrine release. Nat Commun 11:6157
- Heo S, Diering GH, Na CH, Nirujogi RS, Bachman JL, Pandey A, Huganir RL (2018) Identification of long-lived synaptic proteins by proteomic analysis of synaptosome protein turnover. Proc Natl Acad Sci U S A 115:E3827-E3836
- Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, Costantini F (2001) Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol 1:4
- Choi CI, Yoon SP, Choi JM, Kim SS, Lee YD, Birnbaumer L, Suh-Kim H (2014) Simultaneous deletion of floxed genes mediated by CaMKIIα-Cre in the brain and in male germ cells: application to conditional and conventional disruption of Goα. Exp Mol Med 46:e93
- Choi JM, Kim SS, Choi CI, Cha HL, Oh HH, Ghil S, Lee YD, Birnbaumer L, Suh-Kim H (2016) Development of the main olfactory system and main olfactory epithelium-dependent male mating behavior are altered in Go-deficient mice. Proc Natl Acad Sci U S A 113:10974-10979
- Park YM, Chun H, Shin JI, Lee CJ (2018) Astrocyte specificity and coverage of hGFAP-CreERT2 [Tg(GFAP-Cre/ERT2)13Kdmc] mouse line in various brain regions. Exp Neurobiol 27:508-525
- Behesti H, Marino S (2009) Cerebellar granule cells: insights into proliferation, differentiation, and role in medulloblastoma pathogenesis. Int J Biochem Cell Biol 41:435-445
- Consalez GG, Goldowitz D, Casoni F, Hawkes R (2021) Origins, development, and compartmentation of the granule cells of the cerebellum. Front Neural Circuits 14:611841
- Corrales JD, Blaess S, Mahoney EM, Joyner AL (2006) The level of Sonic hedgehog signaling regulates the complexity of cerebellar foliation. Development 133:1811-1821
- Cheng FY, Fleming JT, Chiang C (2018) Bergmann glial Sonic hedgehog signaling activity is required for proper cerebellar cortical expansion and architecture. Dev Biol 440:152-166
- Sotelo C (2004) Cellular and genetic regulation of the development of the cerebellar system. Prog Neurobiol 72:295-339
- Machold R, Fishell G (2005) Math1 is expressed in temporally discrete pools of cerebellar rhombic-lip neural progenitors. Neuron 48:17-24
- Consalez GG, Hawkes R (2013) The compartmental restriction of cerebellar interneurons. Front Neural Circuits 6:123
- Leto K, Carletti B, Williams IM, Magrassi L, Rossi F (2006) Different types of cerebellar GABAergic interneurons originate from a common pool of multipotent progenitor cells. J Neurosci 26:11682-11694
- Leto K, Rolando C, Rossi F (2012) The genesis of cerebellar GABAergic neurons: fate potential and specification mechanisms. Front Neuroanat 6:6
- Huang X, Liu J, Ketova T, Fleming JT, Grover VK, Cooper MK, Litingtung Y, Chiang C (2010) Transventricular delivery of Sonic hedgehog is essential to cerebellar ventricular zone development. Proc Natl Acad Sci U S A 107:8422-8427
- Zhang L, Goldman JE (1996) Generation of cerebellar interneurons from dividing progenitors in white matter. Neuron 16:47-54
- Hoshino M, Nakamura S, Mori K, Kawauchi T, Terao M, Nishimura YV, Fukuda A, Fuse T, Matsuo N, Sone M, Watanabe M, Bito H, Terashima T, Wright CV, Kawaguchi Y, Nakao K, Nabeshima Y (2005) Ptf1a, a bHLH transcriptional gene, defines GABAergic neuronal fates in cerebellum. Neuron 47:201-213
- Weisheit G, Gliem M, Endl E, Pfeffer PL, Busslinger M, Schilling K (2006) Postnatal development of the murine cerebellar cortex: formation and early dispersal of basket, stellate and Golgi neurons. Eur J Neurosci 24:466-478
- Fünfschilling U, Reichardt LF (2002) Cre-mediated recombination in rhombic lip derivatives. Genesis 33:160-169
- Aller MI, Jones A, Merlo D, Paterlini M, Meyer AH, Amtmann U, Brickley S, Jolin HE, McKenzie AN, Monyer H, Farrant M, Wisden W (2003) Cerebellar granule cell Cre recombinase expression. Genesis 36:97-103
- Kim E, Wang Y, Kim SJ, Bornhorst M, Jecrois ES, Anthony TE, Wang C, Li YE, Guan JL, Murphy GG, Zhu Y (2014) Transient inhibition of the ERK pathway prevents cerebellar developmental defects and improves long-term motor functions in murine models of neurofibromatosis type 1. Elife 3:e05151
- Men Y, Zhang A, Li H, Jin Y, Sun X, Li H, Gao J (2015) LKB1 regulates cerebellar development by controlling sonic hedgehog-mediated granule cell precursor proliferation and granule cell migration. Sci Rep 5:16232
- Barski JJ, Dethleffsen K, Meyer M (2000) Cre recombinase expression in cerebellar Purkinje cells. Genesis 28:93-98
- Driver AM, Shumrick C, Stottmann RW (2017) Ttc21b is required in bergmann glia for proper granule cell radial migration. J Dev Biol 5:18
- Sługocka A, Wiaderkiewicz J, Barski JJ (2017) Genetic targeting in cerebellar purkinje cells: an update. Cerebellum 16:191-202
- Pohlkamp T, Steller L, May P, Günther T, Schüle R, Frotscher M, Herz J, Bock HH (2014) Generation and characterization of an Nse-CreERT2 transgenic line suitable for inducible gene manipulation in cerebellar granule cells. PLoS One 9:e100384
- Chow LM, Zhang J, Baker SJ (2008) Inducible Cre recombinase activity in mouse mature astrocytes and adult neural precursor cells. Transgenic Res 17:919-928
- Parmigiani E, Leto K, Rolando C, Figueres-Oñate M, López-Mascaraque L, Buffo A, Rossi F (2015) Heterogeneity and bipotency of astroglial-like cerebellar progenitors along the interneuron and glial lineages. J Neurosci 35:7388-7402
- Guo Z, Wang X, Xiao J, Wang Y, Lu H, Teng J, Wang W (2013) Early postnatal GFAP-expressing cells produce multilineage progeny in cerebrum and astrocytes in cerebellum of adult mice. Brain Res 1532:14-20
- Mori T, Tanaka K, Buffo A, Wurst W, Kühn R, Götz M (2006) Inducible gene deletion in astroglia and radial glia--a valuable tool for functional and lineage analysis. Glia 54:21-34
- Zhang RS, Liakath-Ali K, Südhof TC (2020) Latrophilin-2 and latrophilin-3 are redundantly essential for parallel-fiber synapse function in cerebellum. Elife 9:e54443
- Yang H, Zhu Q, Cheng J, Wu Y, Fan M, Zhang J, Wu H (2019) Opposite regulation of Wnt/β-catenin and Shh signaling pathways by Rack1 controls mammalian cerebellar development. Proc Natl Acad Sci U S A 116:4661-4670
- Koziol LF, Budding D, Andreasen N, D'Arrigo S, Bulgheroni S, Imamizu H, Ito M, Manto M, Marvel C, Parker K, Pezzulo G, Ramnani N, Riva D, Schmahmann J, Vandervert L, Yamazaki T (2014) Consensus paper: the cerebellum's role in movement and cognition. Cerebellum 13:151-177
- Lawrenson C, Bares M, Kamondi A, Kovács A, Lumb B, Apps R, Filip P, Manto M (2018) The mystery of the cerebellum: clues from experimental and clinical observations. Cerebellum Ataxias 5:8
- Massey PV, Bashir ZI (2007) Long-term depression: multiple forms and implications for brain function. Trends Neurosci 30:176-184
- Hirano T (2013) Long-term depression and other synaptic plasticity in the cerebellum. Proc Jpn Acad Ser B Phys Biol Sci 89:183-195
- Hoxha E, Tempia F, Lippiello P, Miniaci MC (2016) Modulation, plasticity and pathophysiology of the parallel fiber-purkinje cell synapse. Front Synaptic Neurosci 8:35





