Articles
Article Tools
Stats or Metrics
Article
Short Communication
Exp Neurobiol 2020; 29(5): 334-343
Published online October 31, 2020
https://doi.org/10.5607/en20049
© The Korean Society for Brain and Neural Sciences
Rho Guanine Nucleotide Exchange Factor 4 (Arhgef4) Deficiency Enhances Spatial and Object Recognition Memory
Ki-Seo Yoo1, Kina Lee1, Yong-Seok Lee2, Won-Jong Oh3 and Hyong Kyu Kim1*
1Department of Medicine and Microbiology, Graduate Program in Neuroscience, College of Medicine, Chungbuk National University, Cheongju 28644, 2Department of Physiology, Department of Biomedical Science, Seoul National University College of Medicine, Seoul 03080, 3Neurovascular Unit Research Group, Korea Brain Research Institute, Daegu 41062, Korea
Correspondence to: *To whom correspondence should be addressed.
TEL: 82-43-261-2867, FAX: 82-43-272-1603
e-mail: hkkim69@chungbuk.ac.kr
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/4.0) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Abstract
Guanine nucleotide exchange factors (GEFs) play multiple functional roles in neurons. In a previous study, we reported that
Graphical Abstract
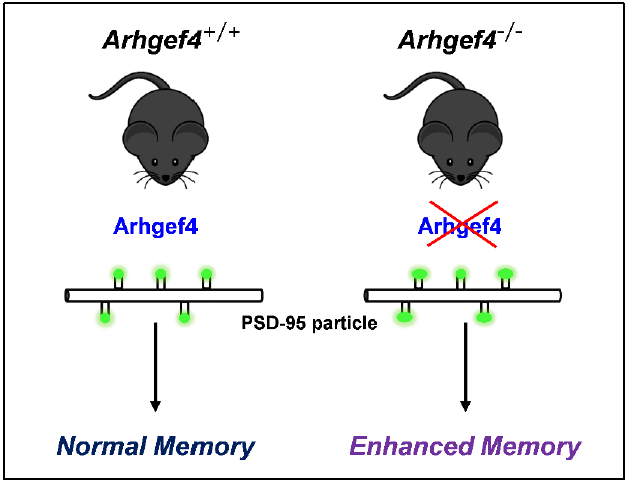
Keywords: Arhgef4, PSD-95, Spatial memory, Recognition
INTRODUCTION
Rho guanine nucleotide exchange factors (GEFs) are involved in the activation of Rho family GTPases by accelerating the exchange of GDP to GTP. Moreover, due to their multiple domains, GEFs act as functional and structural regulators within the postsynaptic regions of neurons in response to external stimuli [1, 2]. Thus, GEFs play a crucial role in various behaviors, such as anxiety, learning, and memory in experimental animals and also in human pathological conditions [2]. For example, the genetic deletion of Kalirin7, a GEF of excitatory synapses for Rac1 and RhoG, shows normal object recognition but impaired passive avoidance fear memory in Kalirin7 knockout (KO) mice [3]. The lack of collybistin, a GEF of inhibitory synapses selectively activating the small GTPase Cdc42, results in a reduced capability of spatial learning and enhances anxiety-like behavior in collybistin-deficient mice [4].
MATERIALS AND METHODS
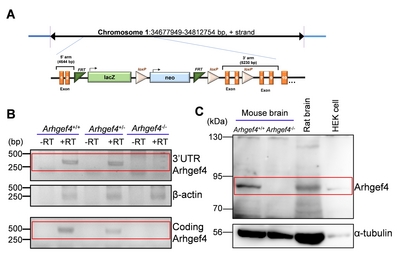
Transgenic animals
RT-PCR and western blotting
Quantitative real-time PCR
The 0.5 μg of total RNAs from the brain of WT, Hetero, or Homo was synthetized to cDNAs, and subsequently used to PCR containing SYBR Green ready mix (TOPrealTM One-step RT qPCR Kit, Enzynomics, Daejeon, Korea) and primers (identical sets used for RT-PCR analysis) by real-time PCR system (CFX96 Touch Real-Time PCR Detection System, Bio-Rad, Laboratory, Hercules, CA, USA). The relative change of
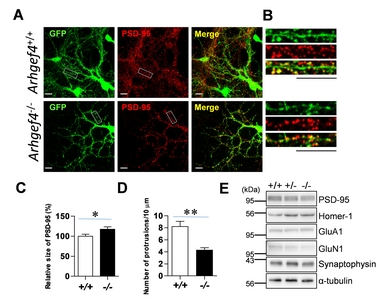
Neuronal culture, immunostaining, and image analysis
Hippocampi were isolated from the brain of postnatal day one (P1) animals and used for culture as previously described [16]. After twelve days, the cultures were infected with Sindbis virus encoding green fluorescent protein (GFP) [11] for 12 h, followed by immunostaining with monoclonal anti-PSD-95 antibody (Clone 6G6-1C9, Thermo Fisher Scientific) and Cy3-conjugated goat anti-mouse IgG (Jackson ImmunoResearch Labs, West Grove, PA, USA). Fluorescent images were acquired with confocal microscopy (Zeiss LSM 800 Airyscan, Carl Zeiss Microscopy GmbH, Jena, Germany), and the acquired images were analyzed with the ImageJ program (ver 1.46r, NIH, Bethesda, MA, USA). Image acquisition and analysis were performed in blinded experiments and image analysis was performed as previously described [16]. The data are presented as mean±standard error of the mean (SEM). The Student’s unpaired
Behavioral tests
All experiments using mice were performed in accordance with the approved animal protocols and the guidelines of the Institutional Animal Care and Use Committee of the Chungbuk National University (CBNUA-1236-19-01). Fewer than four mice were placed in cages on a reversed light-dark cycle and were permitted food and water
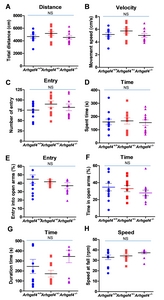
Open field test
Open field tests were performed as described [17]. Tests were performed in an opaque white plastic arena (33×33 cm, 33 cm high). Mice were placed in the periphery of the arena, and their behavior was recorded for 15 min using a camcorder (HDR-CX100, SONY, Tokyo, Japan). For the measurement of general motor activity, path length (total distance) and speed of movement in the total area were analyzed by Ethovision XT (Noldus, Wageningen, the Netherlands). For anxiety-related behavior, entries to the central area and times spent in the central area (infield, square 20×20 cm) were analyzed.
Elevated plus-maze test
Mice were placed in the center of an elevated plus-maze (4×30 cm arms, 60 cm above floor level, 18 cm high non-transparent side walls), and their paths were recorded by a camcorder (HDR-CX100, SONY, Tokyo, Japan). Time spent in each arm and entries into each arm over 10 min were manually scored and changed to percentage. More details are described in a previous study [18].
Rotarod test
Rotarod tests to measure motor skills of mice were performed as described [19]. Mice were placed on the rotating rod with a start speed of 4 rpm, acceleration rate 20 rpm/min (47600, Ugo Basile, Gemonio VA, Italy) and tested for 14 min. Three times trials each 14 min with 15 min interval were performed. Duration time on the rod before mouse falls off and rod spin speed (rapid per minute, rpm) when mouse falls off were scored and averaged.
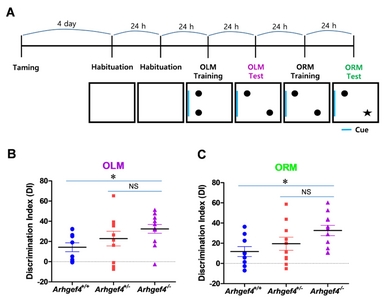
Object location memory test
The OLM test was performed as previously described [20, 21] and included training and test sessions. Before training, mice were habituated for 5 min per day for 4 days in an arena (33 cm×33 cm, 33 cm high, less than 45 LUX) and then habituated for 15 min per day over the next 2 days. One side of the experimental box included a spatial cue. In the training session, mice were allowed to freely explore two identical objects placed in the box for 10 min. During the test session 24 h after the training, mice were placed back in the same box, but one of the objects was moved to a new location. Interaction with each object (defined as sniffing and/or head within 1 cm of the object) was manually scored for analysis. Mice that showed more than 10% preference for each object in the training session were excluded from the subsequent memory tests. The discrimination index was calculated as follows: (time exploring the novel object – time exploring the old)/(time exploring novel+old)×100.
Novel object recognition memory test
After the OLM tests, mice were placed in the arena for the novel ORM tests with the same objects situated in the same location, and allowed to explore for 10 min. Twenty-four hours later, the mice were placed back into the experimental box containing an old object and a new object in the same locations and allowed to explore for 5 min. Mice that showed more than a 10% preference for each object in the training session were excluded from the subsequent memory tests.
Statistical analyses
Data normality was assessed with the Kolmogorov-Smirnov test, the D’Agostino & Pearson Omnibus normality test, or the Shapiro-Wilk normality test. One-way analysis of variance (ANOVA) was used to compare more than two groups. Post hoc comparisons were conducted using Dunnett’s or Bonferroni’s multiple comparison tests. If the data did not follow a Gaussian distribution, a nonparametric Kruskal-Wallis test was used to compare more than two groups. The Student’s unpaired
RESULTS
Arhgef4-deficient mice exhibited increase of PSD-95 particle size in neurons
To investigate the role of
General motor activity and anxiety-like behavior in Arhgef4 KO mice were not altered
First, we examined the general locomotive activity of
Long-term spatial and recognition memories in Arhgef4 KO mice were enhanced
Given that PSD-95 is a key player in synaptic plasticity, which may underlie learning and memory, we examined the long-term memory of
DISCUSSION
In this study, we demonstrated that deficiency of
Consistent with previous reports [6, 7],
PSD-95, a major scaffolding protein in excitatory synapses interacts with many synaptic proteins including signaling molecules, receptors, and channels, and has a pivotal role in synaptic assembly and function [8, 29]. Moreover, PSD-95 levels at postsynapses in excitatory neurons contribute to a variety of memories in experimental animals [9, 10, 30, 31]. Thus, PSD-95 has been the focus of studies on development and synaptic plasticity. In our data,
Even though there are no effects on viability and mortality, the global
In conclusion,
ACKNOWLEDGEMENTS
This research was supported by grants from the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF-2018R1D1A1B07043779) to Hyong Kyu Kim and from the Korea Brain Research Institute (KBRI 20-BR-01-06) to Won-Jong Oh.
FOOTNOTE
Figures
Tables
Fold change of
| Genotype | Average ΔΔCt | Expression fold change to wild type (2-ΔΔCt) |
|---|---|---|
| 0.85±0.20 | 0.555 (0.483~0.637) | |
| 4.29±0.46 | 0.051 (0.037~0.070) |
SD, standard deviation.
References
- Kiraly DD, Eipper-Mains JE, Mains RE, Eipper BA (2010) Synaptic plasticity, a symphony in GEF. ACS Chem Neurosci 1:348-365
- Miller MB, Yan Y, Eipper BA, Mains RE (2013) Neuronal Rho GEFs in synaptic physiology and behavior. Neuroscientist 19:255-273
- Ma XM, Kiraly DD, Gaier ED, Wang Y, Kim EJ, Levine ES, Eipper BA, Mains RE (2008) Kalirin-7 is required for synaptic structure and function. J Neurosci 28:12368-12382
- Papadopoulos T, Korte M, Eulenburg V, Kubota H, Retiounskaia M, Harvey RJ, Harvey K, O'Sullivan GA, Laube B, Hülsmann S, Geiger JR, Betz H (2007) Impaired GABAergic transmission and altered hippocampal synaptic plasticity in collybistin-deficient mice. EMBO J 26:3888-3899
- Kawasaki Y, Senda T, Ishidate T, Koyama R, Morishita T, Iwayama Y, Higuchi O, Akiyama T (2000) Asef, a link between the tumor suppressor APC and G-protein signaling. Science 289:1194-1197
- Hamann MJ, Lubking CM, Luchini DN, Billadeau DD (2007) Asef2 functions as a Cdc42 exchange factor and is stimulated by the release of an autoinhibitory module from a concealed C-terminal activation element. Mol Cell Biol 27:1380-1393
- Thiesen S, Kübart S, Ropers HH, Nothwang HG (2000) Isolation of two novel human RhoGEFs, ARHGEF3 and ARHGEF4, in 3p13-21 and 2q22. Biochem Biophys Res Commun 273:364-369
- Sheng M, Kim E (2011) The postsynaptic organization of synapses. Cold Spring Harb Perspect Biol 3:a005678
- Fitzgerald PJ, Pinard CR, Camp MC, Feyder M, Sah A, Bergstrom HC, Graybeal C, Liu Y, Schlüter OM, Grant SG, Singewald N, Xu W, Holmes A (2015) Durable fear memories require PSD-95. Mol Psychiatry 20:913
- Nithianantharajah J, Komiyama NH, McKechanie A, Johnstone M, Blackwood DH, St Clair D, Emes RD, van de Lagemaat LN, Saksida LM, Bussey TJ, Grant SG (2013) Synaptic scaffold evolution generated components of vertebrate cognitive complexity. Nat Neurosci 16:16-24
- Oh JY, Lim CS, Yoo KS, Park H, Park YS, Kim EG, Lee YS, Kaang BK, Kim HK (2018) Adenomatous polyposis coli-stimulated GEF 1 (Asef1) is a negative regulator of excitatory synaptic function. J Neurochem 147:595-608
- Skarnes WC, Rosen B, West AP, Koutsourakis M, Bushell W, Iyer V, Mujica AO, Thomas M, Harrow J, Cox T, Jackson D, Severin J, Biggs P, Fu J, Nefedov M, de Jong PJ, Stewart AF, Bradley A (2011) A conditional knockout resource for the genome-wide study of mouse gene function. Nature 474:337-342
- Mouse Genome Informatics (2020) Diagrams of general structure of IKMC primary alleles and derivative alleles [Internet]. The Jackson Laboratory, Bar Harbor.
Available from: http://www.informatics.jax.org/mgihome/nomen/IKMC_schematics.shtml - Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25:402-408
- Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:e45
- Yoo KS, Lee K, Oh JY, Lee H, Park H, Park YS, Kim HK (2019) Postsynaptic density protein 95 (PSD-95) is transported by KIF5 to dendritic regions. Mol Brain 12:97
- Bailey KR, Crawley JN (2009) Anxiety-related behaviors in mice. In: Methods of behavioral analysis in neuroscience (Buccafusco JJ ed). 2nd ed. CRC Press, Boca Raton, FL
- Komada M, Takao K, Miyakawa T (2008) Elevated plus maze for mice. J Vis Exp 22:1088
- Deacon RM (2013) Measuring motor coordination in mice. J Vis Exp 75:e2609
- Vogel-Ciernia A, Wood MA (2014) Examining object location and object recognition memory in mice. Curr Protoc Neurosci 69:8.31.1-8.31.17
- Lee YS, Ehninger D, Zhou M, Oh JY, Kang M, Kwak C, Ryu HH, Butz D, Araki T, Cai Y, Balaji J, Sano Y, Nam CI, Kim HK, Kaang BK, Burger C, Neel BG, Silva AJ (2014) Mechanism and treatment for learning and memory deficits in mouse models of Noonan syndrome. Nat Neurosci 17:1736-1743
- International Mouse Phenotyping Consortium (2020) Gene: Arhgef4 [Internet]. International Mouse Phenotyping Consortium.
Available from: https://www.mousephenotype.org/data/genes/MGI:2442507 - Xu W (2011) PSD-95-like membrane associated guanylate kinases (PSD-MAGUKs) and synaptic plasticity. Curr Opin Neurobiol 21:306-312
- El-Husseini AE, Schnell E, Chetkovich DM, Nicoll RA, Bredt DS (2000) PSD-95 involvement in maturation of excitatory synapses. Science 290:1364-1368
- Vogel-Ciernia A, Matheos DP, Barrett RM, Kramár EA, Azzawi S, Chen Y, Magnan CN, Zeller M, Sylvain A, Haettig J, Jia Y, Tran A, Dang R, Post RJ, Chabrier M, Babayan AH, Wu JI, Crabtree GR, Baldi P, Baram TZ, Lynch G, Wood MA (2013) The neuron-specific chromatin regulatory subunit BAF53b is necessary for synaptic plasticity and memory. Nat Neurosci 16:552-561
- Barrett RM, Malvaez M, Kramar E, Matheos DP, Arrizon A, Cabrera SM, Lynch G, Greene RW, Wood MA (2011) Hippocampal focal knockout of CBP affects specific histone modifications, long-term potentiation, and long-term memory. Neuropsychopharmacology 36:1545-1556
- Haettig J, Stefanko DP, Multani ML, Figueroa DX, McQuown SC, Wood MA (2011) HDAC inhibition modulates hippocampus-dependent long-term memory for object location in a CBP-dependent manner. Learn Mem 18:71-79
- The Human Protein Atlas (2020) ARHGEF4 [Internet]. The Human Protein Atlas.
Available from: https://www.proteinatlas.org/ENSG00000136002-ARHGEF4/tissue - Won S, Levy JM, Nicoll RA, Roche KW (2017) MAGUKs: multifaceted synaptic organizers. Curr Opin Neurobiol 43:94-101
- Migaud M, Charlesworth P, Dempster M, Webster LC, Watabe AM, Makhinson M, He Y, Ramsay MF, Morris RG, Morrison JH, O'Dell TJ, Grant SG (1998) Enhanced long-term potentiation and impaired learning in mice with mutant postsynaptic density-95 protein. Nature 396:433-439
- Elkobi A, Ehrlich I, Belelovsky K, Barki-Harrington L, Rosenblum K (2008) ERK-dependent PSD-95 induction in the gustatory cortex is necessary for taste learning, but not retrieval. Nat Neurosci 11:1149-1151





